5XJ8
 
 | | Crystal structure of PlsY (YgiH), an integral membrane glycerol 3-phosphate acyltransferase - the lysphosphatidic acid form | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl hexadecanoate, Glycerol-3-phosphate acyltransferase, PHOSPHATE ION | | Authors: | Li, Z, Tang, Y, Li, D. | | Deposit date: | 2017-04-30 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural insights into the committed step of bacterial phospholipid biosynthesis.
Nat Commun, 8, 2017
|
|
4WAB
 
 | | Crystal structure of mPGES1 solved by native-SAD phasing | | Descriptor: | 2-[[2,6-bis(chloranyl)-3-[(2,2-dimethylpropanoylamino)methyl]phenyl]amino]-1-methyl-6-(2-methyl-2-oxidanyl-propoxy)-N-[2,2,2-tris(fluoranyl)ethyl]benzimidazole-5-carboxamide, GLUTATHIONE, Prostaglandin E synthase,Leukotriene C4 synthase | | Authors: | Weinert, T, Li, D, Howe, N, Caffrey, M, Wang, M. | | Deposit date: | 2014-08-29 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
7F5G
 
 | | The crystal structure of RBD-Nanobody complex, DL4 (SA4) | | Descriptor: | ACETATE ION, GLYCEROL, Nanobody DL4, ... | | Authors: | Li, T, Lai, Y, Zhou, Y, Tan, J, Li, D. | | Deposit date: | 2021-06-22 | | Release date: | 2022-05-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Isolation, characterization, and structure-based engineering of a neutralizing nanobody against SARS-CoV-2.
Int.J.Biol.Macromol., 209, 2022
|
|
7F5H
 
 | | The crystal structure of RBD-Nanobody complex, DL28 (SC4) | | Descriptor: | GLYCEROL, Nanobody DL28, PHOSPHATE ION, ... | | Authors: | Luo, Z.P, Li, T, Lai, Y, Zhou, Y, Tan, J, Li, D. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Characterization of a Neutralizing Nanobody With Broad Activity Against SARS-CoV-2 Variants.
Front Microbiol, 13, 2022
|
|
8I23
 
 | | Clostridium thermocellum RNA polymerase transcription open complex with SigI1 and its promoter | | Descriptor: | DNA (80-mer), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Li, J, Zhang, H, Li, D, Feng, Y, Zhu, P. | | Deposit date: | 2023-01-13 | | Release date: | 2023-10-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structure of the transcription open complex of distinct sigma I factors.
Nat Commun, 14, 2023
|
|
8I24
 
 | | Clostridium thermocellum RNA polymerase transcription open complex with SigI6 and its promoter | | Descriptor: | DNA (80-mer), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Li, J, Zhang, H, Li, D, Feng, Y, Zhu, P. | | Deposit date: | 2023-01-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structure of the transcription open complex of distinct sigma I factors.
Nat Commun, 14, 2023
|
|
8IY7
 
 | |
8IZZ
 
 | |
8HZC
 
 | |
8HZB
 
 | |
8HZS
 
 | |
8HOY
 
 | | Cryo-EM structure of monkeypox virus DNA replication holoenzyme F8, A22 and E4 complex without DNA at 2.76 angostram | | Descriptor: | DNA polymerase, DNA polymerase processivity factor component A20, E4R | | Authors: | Xu, Y, Wu, Y, Zhang, Y, Fan, R, Yang, Y, Li, D, Yang, B, Zhang, Z, Dong, C. | | Deposit date: | 2022-12-11 | | Release date: | 2023-12-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Cryo-EM structures of human monkeypox viral replication complexes with and without DNA duplex.
Cell Res., 33, 2023
|
|
8HPA
 
 | | Monkeypox virus DNA replication holoenzyme F8, A22 and E4 complex in a DNA binding form | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*CP*TP*TP*CP*CP*CP*CP*TP*AP*C)-3'), DNA (5'-D(P*AP*TP*GP*GP*TP*AP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*CP*G)-3'), DNA polymerase, ... | | Authors: | Xu, Y, Wu, Y, Zhang, Y, Fan, R, Yang, Y, Li, D, Yang, B, Zhang, Z, Dong, C. | | Deposit date: | 2022-12-12 | | Release date: | 2024-01-31 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Cryo-EM structures of human monkeypox viral replication complexes with and without DNA duplex.
Cell Res., 33, 2023
|
|
4ZWJ
 
 | | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser | | Descriptor: | Chimera protein of human Rhodopsin, mouse S-arrestin, and T4 Endolysin | | Authors: | Kang, Y, Zhou, X.E, Gao, X, He, Y, Liu, W, Ishchenko, A, Barty, A, White, T.A, Yefanov, O, Han, G.W, Xu, Q, de Waal, P.W, Ke, J, Tan, M.H.E, Zhang, C, Moeller, A, West, G.M, Pascal, B, Eps, N.V, Caro, L.N, Vishnivetskiy, S.A, Lee, R.J, Suino-Powell, K.M, Gu, X, Pal, K, Ma, J, Zhi, X, Boutet, S, Williams, G.J, Messerschmidt, M, Gati, C, Zatsepin, N.A, Wang, D, James, D, Basu, S, Roy-Chowdhury, S, Conrad, C, Coe, J, Liu, H, Lisova, S, Kupitz, C, Grotjohann, I, Fromme, R, Jiang, Y, Tan, M, Yang, H, Li, J, Wang, M, Zheng, Z, Li, D, Howe, N, Zhao, Y, Standfuss, J, Diederichs, K, Dong, Y, Potter, C.S, Carragher, B, Caffrey, M, Jiang, H, Chapman, H.N, Spence, J.C.H, Fromme, P, Weierstall, U, Ernst, O.P, Katritch, V, Gurevich, V.V, Griffin, P.R, Hubbell, W.L, Stevens, R.C, Cherezov, V, Melcher, K, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2015-05-19 | | Release date: | 2015-07-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.302 Å) | | Cite: | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser.
Nature, 523, 2015
|
|
5DWK
 
 | | Diacylglycerol Kinase solved by multi crystal multi orientation native SAD | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, ACETATE ION, ... | | Authors: | Weinert, T, Olieric, V, Finke, A.D, Li, D, Caffrey, M, Wang, M. | | Deposit date: | 2015-09-22 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Data-collection strategy for challenging native SAD phasing.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6LP2
 
 | | Structure of Lpg2148/UBE2N-Ub complex | | Descriptor: | Ubiquitin, Ubiquitin-conjugating enzyme E2 N, Uncharacterized protein lpg2148 | | Authors: | Feng, Y, Wang, Y, Huang, Y, Li, D. | | Deposit date: | 2020-01-08 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.479 Å) | | Cite: | Structure of Lpg2148/UBE2N-Ub complex
To Be Published
|
|
6LZ2
 
 | | Crystal structure of a thermostable green fluorescent protein (TGP) with a synthetic nanobody (Sb44) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Cai, H, Yao, H, Li, T, Hutter, C, Tang, Y, Li, Y, Seeger, M, Li, D. | | Deposit date: | 2020-02-17 | | Release date: | 2020-12-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | An improved fluorescent tag and its nanobodies for membrane protein expression, stability assay, and purification.
Commun Biol, 3, 2020
|
|
5M5Z
 
 | | Chaetomium thermophilum beta-1-3-glucanase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-1,3-glucanase, ... | | Authors: | Papageorgiou, A.C, Chen, J, Li, D. | | Deposit date: | 2016-10-23 | | Release date: | 2017-05-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure and biological implications of a glycoside hydrolase family 55 beta-1,3-glucanase from Chaetomium thermophilum.
Biochim. Biophys. Acta, 1865, 2017
|
|
5M60
 
 | | Chaetomium thermophilum beta-1-3-glucanase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-1,3-glucanase, SODIUM ION, ... | | Authors: | Papageorgiou, A.C, Chen, J, Li, D. | | Deposit date: | 2016-10-23 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure and biological implications of a glycoside hydrolase family 55 beta-1,3-glucanase from Chaetomium thermophilum.
Biochim. Biophys. Acta, 1865, 2017
|
|
2RI1
 
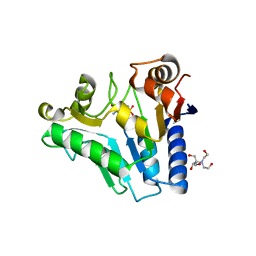 | | Crystal Structure of glucosamine 6-phosphate deaminase (NagB) with GlcN6P from S. mutans | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, Glucosamine-6-phosphate deaminase | | Authors: | Liu, C, Li, D, Su, X.D. | | Deposit date: | 2007-10-10 | | Release date: | 2008-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Ring-opening mechanism revealed by crystal structures of NagB and its ES intermediate complex
J.Mol.Biol., 379, 2008
|
|
6A6B
 
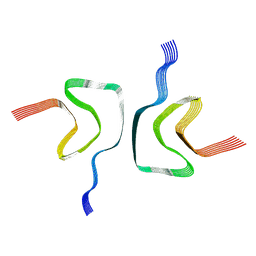 | | cryo-em structure of alpha-synuclein fiber | | Descriptor: | Alpha-synuclein | | Authors: | Li, Y.W, Zhao, C.Y, Luo, F, Liu, Z, Gui, X, Luo, Z, Zhang, X, Li, D, Liu, C, Li, X. | | Deposit date: | 2018-06-27 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Amyloid fibril structure of alpha-synuclein determined by cryo-electron microscopy
Cell Res., 28, 2018
|
|
9J23
 
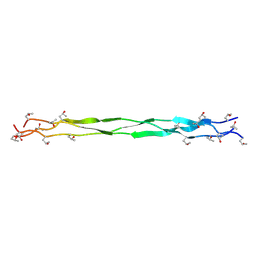 | | Structure of a triple-helix region of human Collagen type II from Trautec | | Descriptor: | Triple-helix region of human collagen type II | | Authors: | Fan, X, Zhai, Y, Chu, Y, Fu, S, Li, D, Feng, P, Cao, K, Li, J, Si, Y, Ma, L, Qian, S. | | Deposit date: | 2024-08-06 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of a triple-helix region of human Collagen type II from Trautec
To Be Published
|
|
5ZO2
 
 | | Crystal structure of mouse nectin-like molecule 4 (mNecl-4) full ectodomain in complex with mouse nectin-like molecule 1 (mNecl-1) Ig1 domain, 3.3A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cell adhesion molecule 3, Cell adhesion molecule 4 | | Authors: | Liu, X, An, T, Li, D, Fan, Z, Xiang, P, Li, C, Ju, W, Li, J, Hu, G, Qin, B, Yin, B, Wojdyla, J.A, Wang, M, Yuan, J, Qiang, B, Shu, P, Cui, S, Peng, X. | | Deposit date: | 2018-04-12 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structure of the heterophilic interaction between the nectin-like 4 and nectin-like 1 molecules.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5ZO1
 
 | | Crystal structure of mouse nectin-like molecule 4 (mNecl-4) full ectodomain (Ig1-Ig3), 2.2A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cell adhesion molecule 4, GLYCEROL | | Authors: | Liu, X, An, T, Li, D, Fan, Z, Xiang, P, Li, C, Ju, W, Li, J, Hu, G, Qin, B, Yin, B, Wojdyla, J.A, Wang, M, Yuan, J, Qiang, B, Shu, P, Cui, S, Peng, X. | | Deposit date: | 2018-04-12 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structure of the heterophilic interaction between the nectin-like 4 and nectin-like 1 molecules.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
7CYV
 
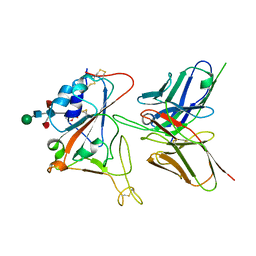 | | Crystal structure of FD20, a neutralizing single-chain variable fragment (scFv) in complex with SARS-CoV-2 Spike receptor-binding domain (RBD) | | Descriptor: | Spike protein S1, The heavy chain variable region of the scFv FD20,The light chain variable region of the scFv FD20, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)][alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Li, Y, Li, T, Lai, Y, Cai, H, Yao, H, Li, D. | | Deposit date: | 2020-09-04 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Uncovering a conserved vulnerability site in SARS-CoV-2 by a human antibody.
Embo Mol Med, 13, 2021
|
|
