8XD7
 
 | |
8XDF
 
 | |
8XDC
 
 | | Cryo-EM structure of human urea transporter A2. | | Descriptor: | N-[3-[1,1-bis(oxidanylidene)-1,2-thiazolidin-2-yl]-4-chloranyl-phenyl]-2-methoxy-5-methyl-benzenesulfonamide, Urea transporter 2 | | Authors: | Huang, S, Liu, L, Sun, J. | | Deposit date: | 2023-12-10 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the mechanisms of urea permeation and distinct inhibition modes of urea transporters.
Nat Commun, 15, 2024
|
|
8XDI
 
 | | Cryo-EM structure of zebrafish urea transporter. | | Descriptor: | N-(4-acetamidophenyl)-5-ethanoyl-furan-2-carboxamide, Urea transporter | | Authors: | Huang, S, Liu, L, Sun, J. | | Deposit date: | 2023-12-11 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the mechanisms of urea permeation and distinct inhibition modes of urea transporters.
Nat Commun, 15, 2024
|
|
9B4U
 
 | | Crystal structure of p110alpha-RBD covalently bound to a breaker compound BBO-10203 via Cys242 | | Descriptor: | 1-[(4R,8R)-2-[(4M,7P)-7-[2,4-difluoro-6-(2-methoxyethoxy)phenyl]-4-(1-methyl-1H-indazol-5-yl)thieno[3,2-c]pyridin-6-yl]-4-methyl-6,7-dihydropyrazolo[1,5-a]pyrazin-5(4H)-yl]propan-1-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Czyzyk, D.J, Simanshu, D.K. | | Deposit date: | 2024-03-21 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | BBO-10203 inhibits tumor growth without inducing hyperglycemia by blocking RAS-PI3K alpha interaction.
Science, 2025
|
|
7SHU
 
 | | IgE-Fc in complex with omalizumab variant C02 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin heavy constant epsilon, ... | | Authors: | Pennington, L.F, Jardetzky, T.J, Kleinboelting, S. | | Deposit date: | 2021-10-11 | | Release date: | 2021-12-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Directed evolution of and structural insights into antibody-mediated disruption of a stable receptor-ligand complex.
Nat Commun, 12, 2021
|
|
7SHY
 
 | | IgE-Fc in complex with omalizumab scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, IgE Fc, ... | | Authors: | Pennington, L.F, Jardetzky, T.J, Kleinboelting, S. | | Deposit date: | 2021-10-11 | | Release date: | 2021-12-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Directed evolution of and structural insights into antibody-mediated disruption of a stable receptor-ligand complex.
Nat Commun, 12, 2021
|
|
7SI0
 
 | | IgE-Fc in complex with 813 | | Descriptor: | 813 Variable fragment Heavy chain, 813 Variable fragment Light chain, IgE Fc, ... | | Authors: | Pennington, L.F, Jardetzky, T.J, Kleinboelting, S. | | Deposit date: | 2021-10-12 | | Release date: | 2021-12-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Directed evolution of and structural insights into antibody-mediated disruption of a stable receptor-ligand complex.
Nat Commun, 12, 2021
|
|
7SHT
 
 | |
7SHZ
 
 | | IgE-Fc in complex with HAE | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Pennington, L.F, Jardetzky, T.J, Kleinboelting, S. | | Deposit date: | 2021-10-11 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Directed evolution of and structural insights into antibody-mediated disruption of a stable receptor-ligand complex.
Nat Commun, 12, 2021
|
|
7M8W
 
 | | XFEL crystal structure of the prostaglandin D2 receptor CRTH2 in complex with 15R-methyl-PGD2 | | Descriptor: | 15R-methyl-prostaglandin D2, CITRATE ANION, Prostaglandin D2 receptor 2, ... | | Authors: | Shiriaeva, A, Han, G.W, Cherezov, V. | | Deposit date: | 2021-03-30 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Molecular basis for lipid recognition by the prostaglandin D 2 receptor CRTH2.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6OT0
 
 | | Structure of human Smoothened-Gi complex | | Descriptor: | 17-[3-(3,3-DIMETHYL-OXIRANYL)-1-METHYL-PROPYL]-10,13-DIMETHYL-2,3,4,7,8,9,10,11,12,13,14,15,16,17-TETRADECAHYDRO-1H-CYC LOPENTA[A]PHENANTHREN-3-OL, Fab heavy chain, Fab light chain, ... | | Authors: | Qi, X, Li, X. | | Deposit date: | 2019-05-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Cryo-EM structure of oxysterol-bound human Smoothened coupled to a heterotrimeric Gi.
Nature, 571, 2019
|
|
6SCF
 
 | | A viral anti-CRISPR subverts type III CRISPR immunity by rapid degradation of cyclic oligoadenylate | | Descriptor: | Uncharacterized protein, cyclic oligoadenylate | | Authors: | McMahon, S.A, Athukoralage, J.S, Graham, S, White, M.F, Gloster, T.M. | | Deposit date: | 2019-07-24 | | Release date: | 2019-10-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | An anti-CRISPR viral ring nuclease subverts type III CRISPR immunity.
Nature, 577, 2020
|
|
7WUQ
 
 | | Tethered peptide activation mechanism of adhesion GPCRs ADGRG2 and ADGRG4 | | Descriptor: | Adhesion G-protein coupled receptor G2,mCherry, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Q.T, Guo, S.C, Xiao, P, Sun, J.P, Yu, X, Gou, L, Kong, L.L, Zhang, L. | | Deposit date: | 2022-02-09 | | Release date: | 2022-04-27 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Tethered peptide activation mechanism of the adhesion GPCRs ADGRG2 and ADGRG4.
Nature, 604, 2022
|
|
8XKN
 
 | | Cryo-EM structure of tail tube protein | | Descriptor: | a protein | | Authors: | Zhang, H, Li, Z, Li, X.Z. | | Deposit date: | 2023-12-23 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Insights into the modulation of bacterial NADase activity by phage proteins.
Nat Commun, 15, 2024
|
|
7XKE
 
 | | Cryo-EM structure of DHEA-ADGRG2-FL-Gs complex | | Descriptor: | 3-BETA-HYDROXY-5-ANDROSTEN-17-ONE, Adhesion G-protein coupled receptor G2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Guo, S.C, Xiao, P, Lin, H, Sun, J.P, Yu, X. | | Deposit date: | 2022-04-19 | | Release date: | 2022-08-10 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of the ADGRG2-G s complex in apo and ligand-bound forms.
Nat.Chem.Biol., 18, 2022
|
|
7XKD
 
 | | Cryo-EM structure of DHEA-ADGRG2-BT-Gs complex | | Descriptor: | 3-BETA-HYDROXY-5-ANDROSTEN-17-ONE, Adhesion G-protein coupled receptor G2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Guo, S.C, Xiao, P, Lin, H, Sun, J.P, Yu, X. | | Deposit date: | 2022-04-19 | | Release date: | 2022-08-10 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structures of the ADGRG2-G s complex in apo and ligand-bound forms.
Nat.Chem.Biol., 18, 2022
|
|
7XKF
 
 | | Cryo-EM structure of DHEA-ADGRG2-BT-Gs complex at lower state | | Descriptor: | 3-BETA-HYDROXY-5-ANDROSTEN-17-ONE, Adhesion G-protein coupled receptor G2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Guo, S.C, Xiao, P, Lin, H, Sun, J.P, Yu, X. | | Deposit date: | 2022-04-19 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structures of the ADGRG2-G s complex in apo and ligand-bound forms.
Nat.Chem.Biol., 18, 2022
|
|
8K98
 
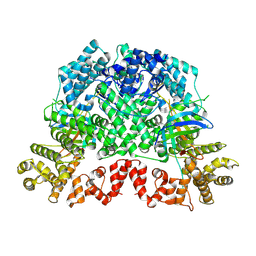 | | Cryo-EM structure of DSR2-TTP | | Descriptor: | a protein | | Authors: | Zhang, H, Li, Z, Li, X.Z. | | Deposit date: | 2023-07-31 | | Release date: | 2024-05-01 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Insights into the modulation of bacterial NADase activity by phage proteins.
Nat Commun, 15, 2024
|
|
8K9A
 
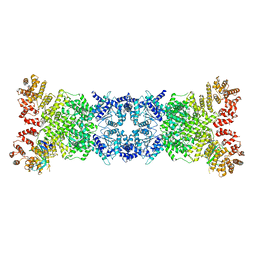 | | Cryo-EM structure of DSR2-DSAD1 state 2 | | Descriptor: | SIR2-like domain-containing protein, SPbeta prophage-derived uncharacterized protein YotI | | Authors: | Zhang, H, Li, Z, Li, X.Z. | | Deposit date: | 2023-07-31 | | Release date: | 2024-05-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Insights into the modulation of bacterial NADase activity by phage proteins.
Nat Commun, 15, 2024
|
|
7Y9V
 
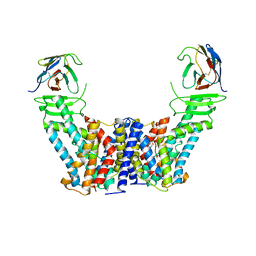 | | Structure of the auxin exporter PIN1 in Arabidopsis thaliana in the IAA-bound state | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, Auxin efflux carrier component 1, nanobody | | Authors: | Sun, L, Liu, X, Yang, Z, Xia, J. | | Deposit date: | 2022-06-26 | | Release date: | 2022-09-07 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into auxin recognition and efflux by Arabidopsis PIN1.
Nature, 609, 2022
|
|
7Y9U
 
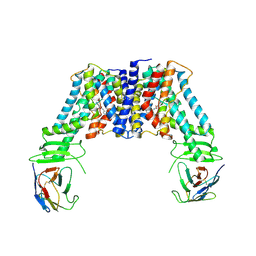 | | Structure of the auxin exporter PIN1 in Arabidopsis thaliana in the NPA-bound state | | Descriptor: | 2-(naphthalen-1-ylcarbamoyl)benzoic acid, Auxin efflux carrier component 1, nanobody | | Authors: | Sun, L, Liu, X, Yang, Z, Xia, J. | | Deposit date: | 2022-06-26 | | Release date: | 2022-09-07 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into auxin recognition and efflux by Arabidopsis PIN1.
Nature, 609, 2022
|
|
7Y9T
 
 | | Structure of the auxin exporter PIN1 in Arabidopsis thaliana in the apo state | | Descriptor: | Auxin efflux carrier component 1, nanobody | | Authors: | Sun, L, Liu, X, Yang, Z, Xia, J. | | Deposit date: | 2022-06-26 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into auxin recognition and efflux by Arabidopsis PIN1.
Nature, 609, 2022
|
|
7VKK
 
 | | Crystal structure of D. melanogaster SAMTOR V66W/E67P mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-adenosylmethionine sensor upstream of mTORC1, SULFATE ION | | Authors: | Zhang, T, Ding, J. | | Deposit date: | 2021-09-30 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Molecular mechanism of S -adenosylmethionine sensing by SAMTOR in mTORC1 signaling.
Sci Adv, 8, 2022
|
|
7VKR
 
 | |
