5FYC
 
 | | Crystal structure of human JMJD2A in complex with succinate | | Descriptor: | 1,2-ETHANEDIOL, FUMARIC ACID, LYSINE-SPECIFIC DEMETHYLASE 4A, ... | | Authors: | Nowak, R, Kopec, J, Johansson, C, Szykowska, A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2016-03-07 | | Release date: | 2016-03-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.261 Å) | | Cite: | Crystal Structure of Human Jmjd2A in Complex with Succinate
To be Published
|
|
8FE5
 
 | | Structure of J-PKAc chimera complexed with Aplithianine B | | Descriptor: | 6-[(6P)-6-(1-methyl-1H-imidazol-5-yl)-2,3-dihydro-4H-1,4-thiazin-4-yl]-7,9-dihydro-8H-purin-8-one, DnaJ homolog subfamily B member 1,cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Du, L, Wilson, B.A.P, Li, N, Martinez Fiesco, J.A, Dalilian, M, Wang, D, Smith, E.A, Wamiru, A, Goncharova, E.I, Zhang, P, O'Keefe, B.R. | | Deposit date: | 2022-12-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery and Synthesis of a Naturally Derived Protein Kinase Inhibitor that Selectively Inhibits Distinct Classes of Serine/Threonine Kinases.
J.Nat.Prod., 86, 2023
|
|
5FT5
 
 | | Crystal structure of the cysteine desulfurase CsdA (persulfurated) from Escherichia coli at 2.384 Angstroem resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Fernandez, F.J, Arda, A, Lopez-Estepa, M, Aranda, J, Penya-Soler, E, Garces, F, Quintana, J.F, Round, A, Campos-Oliva, R, Bruix, M, Coll, M, Tunon, I, Jimenez-Barbero, J, Vega, M.C. | | Deposit date: | 2016-01-11 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.384 Å) | | Cite: | Mechanism of Sulfur Transfer Across Protein-Protein Interfaces: The Cysteine Desulfurase Model System
Acs Catalysis, 6, 2016
|
|
7AMZ
 
 | | Crystal structure of human Butyrylcholinesterase in complex with N-((2S,3R)-4-((2,2-dimethylpropyl)amino)-3-hydroxy-1-phenylbutan-2-yl)-2,2-diphenylacetamide | | Descriptor: | 2,2-dimethylpropyl-[(2~{R},3~{S})-3-(2,2-diphenylethanoylamino)-2-oxidanyl-4-phenyl-butyl]azanium, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Pasieka, A, Panek, D, Wieckowska, A. | | Deposit date: | 2020-10-10 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery of multifunctional anti-Alzheimer's agents with a unique mechanism of action including inhibition of the enzyme butyrylcholinesterase and gamma-aminobutyric acid transporters.
Eur.J.Med.Chem., 218, 2021
|
|
3IHF
 
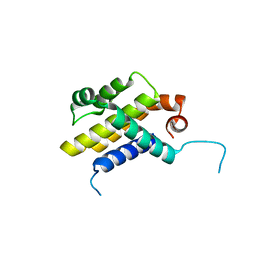 | |
5FT1
 
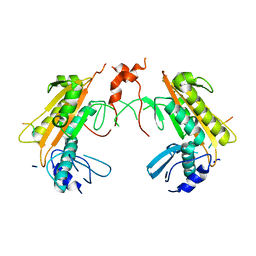 | | Crystal structure of gp37(Dip) from bacteriophage phiKZ bound to RNase E of Pseudomonas aeruginosa | | Descriptor: | GP37, RIBONUCLEASE E | | Authors: | Van den Bossche, A, Hardwick, S.W, Ceyssens, P.J, Hendrix, H, Voet, M, Dendooven, T, Bandyra, K.J, De Maeyer, M, Aertsen, A, Noben, J.P, Luisi, B.F, Lavigne, R. | | Deposit date: | 2016-01-08 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural elucidation of a novel mechanism for the bacteriophage-based inhibition of the RNA degradosome.
Elife, 5, 2016
|
|
6B3S
 
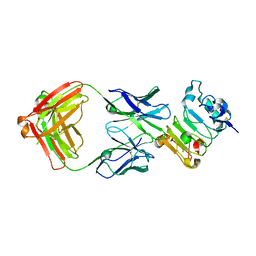 | |
7AV6
 
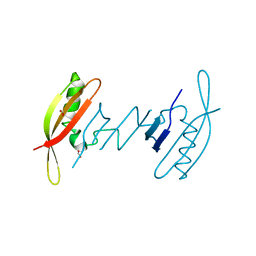 | | FAST in a domain-swapped dimer form | | Descriptor: | FORMIC ACID, Photoactive yellow protein | | Authors: | Bukhdruker, S, Remeeva, A, Ruchkin, D, Gorbachev, D, Povarova, N, Mineev, K, Goncharuk, S, Baranov, M, Mishin, A, Borshchevskiy, V. | | Deposit date: | 2020-11-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | NanoFAST: structure-based design of a small fluorogen-activating protein with only 98 amino acids.
Chem Sci, 12, 2021
|
|
3IIH
 
 | |
7CAT
 
 | | The NADPH binding site on beef liver catalase | | Descriptor: | CATALASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Murthy, M.R.N, Reid III, T.J, Sicignano, A, Tanaka, N, Fita, I, Rossmann, M.G. | | Deposit date: | 1984-11-15 | | Release date: | 1985-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The NADPH binding site on beef liver catalase.
Proc.Natl.Acad.Sci.USA, 82, 1985
|
|
6B7C
 
 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with N-((1,3-dimethyl-1H-pyrazol-5-yl)methyl)-5-methyl-1H-imidazo[4,5-b]pyridin-2-amine | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, N-[(1,3-dimethyl-1H-pyrazol-5-yl)methyl]-5-methyl-3H-imidazo[4,5-b]pyridin-2-amine, ... | | Authors: | Proudfoot, A.W, Bussiere, D, Lingel, A. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.564 Å) | | Cite: | High-Confidence Protein-Ligand Complex Modeling by NMR-Guided Docking Enables Early Hit Optimization.
J. Am. Chem. Soc., 139, 2017
|
|
6B7A
 
 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with 2-methyl-1H-benzo[d]imidazol-4-ol | | Descriptor: | 2-methyl-1H-benzimidazol-7-ol, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Proudfoot, A.W, Bussiere, D, Lingel, A. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | High-Confidence Protein-Ligand Complex Modeling by NMR-Guided Docking Enables Early Hit Optimization.
J. Am. Chem. Soc., 139, 2017
|
|
5J6S
 
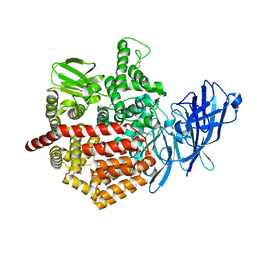 | | Crystal structure of Endoplasmic Reticulum Aminopeptidase 2 (ERAP2) in complex with a hydroxamic derivative ligand | | Descriptor: | (2S)-N~1~-benzyl-2-[(4-fluorophenyl)methyl]-N~3~-hydroxypropanediamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Saridakis, E, Giastas, P, Mpakali, A, Deprez-Poulain, R, Stratikos, E. | | Deposit date: | 2016-04-05 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of ERAP2 Complexed with Inhibitors Reveal Pharmacophore Requirements for Optimizing Inhibitor Potency.
ACS Med Chem Lett, 8, 2017
|
|
7ASX
 
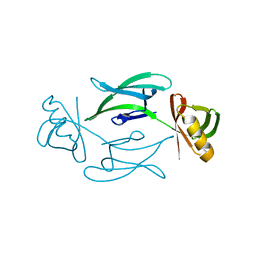 | | Fixed-target serial femtosecond crystallography using in cellulo grown Neurospora crassa HEX-1 microcrystals. (Chip 1) | | Descriptor: | eIF-5a domain-containing protein | | Authors: | Lahey-Rudolph, J.M, Schoenherr, R, Barthelmess, M, Fischer, P, Seuring, C, Wagner, A, Meents, A, Redecke, L. | | Deposit date: | 2020-10-28 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fixed-target serial femtosecond crystallography using in cellulo grown microcrystals.
Iucrj, 8, 2021
|
|
7ASI
 
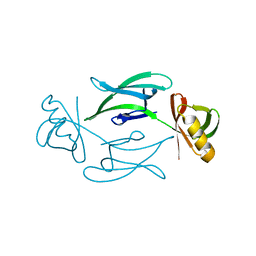 | | Fixed-target serial femtosecond crystallography using in cellulo grown Neurospora crassa HEX-1 microcrystals. (Chips 1+2) | | Descriptor: | eIF-5a domain-containing protein | | Authors: | Lahey-Rudolph, J.M, Schoenherr, R, Barthelmess, M, Fischer, P, Seuring, C, Wagner, A, Meents, A, Redecke, L. | | Deposit date: | 2020-10-27 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | Fixed-target serial femtosecond crystallography using in cellulo grown microcrystals.
Iucrj, 8, 2021
|
|
5FUT
 
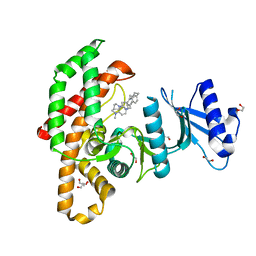 | | Human choline kinase a1 in complex with compound 4-(dimethylamino)-1-{4-[4-(4-{[4-(pyrrolidin- 1-yl)pyridinium-1-yl]methyl}phenyl)butyl]benzyl}pyridinium (compound BR25) | | Descriptor: | 1,2-ETHANEDIOL, 4-(dimethylamino)-1-{4-[4-(4-{[4-(pyrrolidin-1-yl)pyridinium-1-yl]methyl}phenyl)butyl]benzyl}pyridinium, CHOLINE KINASE ALPHA | | Authors: | Serran-Aguilera, L, Denton, H, Rubio-Ruiz, B, Lopez-Gutierrez, B, Entrena, A, Izquierdo, L, Smith, T.K, Conejo-Garcia, A, Hurtado-Guerrero, R. | | Deposit date: | 2016-01-29 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Plasmodium Falciparum Choline Kinase Inhibition Leads to a Major Decrease in Phosphatidylethanolamine Causing Parasite Death.
Sci.Rep., 6, 2016
|
|
5FVA
 
 | | Toscana Virus Nucleocapsid Protein | | Descriptor: | NUCLEOPROTEIN, SODIUM ION | | Authors: | Baklouti, A, Sevajol, M, Goulet, A, Lichiere, J, Ferron, F, Canard, B, Charrel, R.N, Coutard, B, Papageorgiou, N. | | Deposit date: | 2016-02-03 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Toscana virus nucleoprotein oligomer organization observed in solution.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5J94
 
 | | Human cathepsin K mutant C25S in complex with the allosteric effector NSC13345 | | Descriptor: | 2-{[(carbamoylsulfanyl)acetyl]amino}benzoic acid, Cathepsin K, SULFATE ION | | Authors: | Novinec, M, Korenc, M, Lenarcic, B, Baici, A. | | Deposit date: | 2016-04-08 | | Release date: | 2016-04-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.22002459 Å) | | Cite: | A novel allosteric mechanism in the cysteine peptidase cathepsin K discovered by computational methods.
Nat Commun, 5, 2014
|
|
7ZTB
 
 | |
7ZUB
 
 | | Cryo-EM structure of the indirubin-bound Hsp90-XAP2-AHR complex | | Descriptor: | (3~{Z})-3-(3-oxidanylidene-1~{H}-indol-2-ylidene)-1~{H}-indol-2-one, ADENOSINE-5'-DIPHOSPHATE, AH receptor-interacting protein, ... | | Authors: | Gruszczyk, J, Savva, C.G, Lai-Kee-Him, J, Bous, J, Ancelin, A, Kwong, H.S, Grandvuillemin, L, Bourguet, W. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-23 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Cryo-EM structure of the agonist-bound Hsp90-XAP2-AHR cytosolic complex.
Nat Commun, 13, 2022
|
|
5MPK
 
 | | Crystal structure of CREBBP bromodomain complexed with DK19 | | Descriptor: | CREB-binding protein, ~{N}-(5-ethanoyl-2-ethoxy-phenyl)-3-(2~{H}-1,2,3,4-tetrazol-5-yl)-5-(1,3-thiazol-4-yl)benzamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2016-12-16 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
6SGA
 
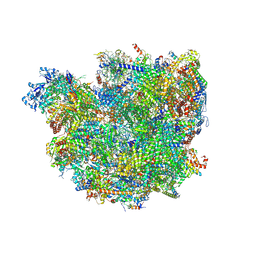 | | Body domain of the mt-SSU assemblosome from Trypanosoma brucei | | Descriptor: | 9S rRNA, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Saurer, M, Ramrath, D.J.F, Niemann, M, Calderaro, S, Prange, C, Mattei, S, Scaiola, A, Leitner, A, Bieri, P, Horn, E.K, Leibundgut, M, Boehringer, D, Schneider, A, Ban, N. | | Deposit date: | 2019-08-03 | | Release date: | 2019-09-18 | | Last modified: | 2019-09-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mitoribosomal small subunit biogenesis in trypanosomes involves an extensive assembly machinery.
Science, 365, 2019
|
|
5FYA
 
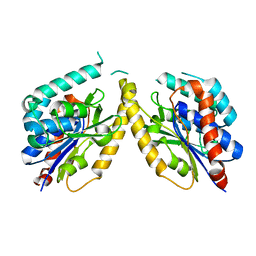 | | Cubic crystal of the native PlpD | | Descriptor: | PATATIN-LIKE PROTEIN, PLPD | | Authors: | Vinicius da Mata Madeira, P, Zouhir, S, Basso, P, Neves, D, Laubier, A, Salacha, R, Bleves, S, Faudry, E, Contreras-Martel, C, Dessen, A. | | Deposit date: | 2016-03-04 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Structural Basis of Lipid Targeting and Destruction by the Type V Secretion System of Pseudomonas Aeruginosa.
J.Mol.Biol., 428, 2016
|
|
3B8N
 
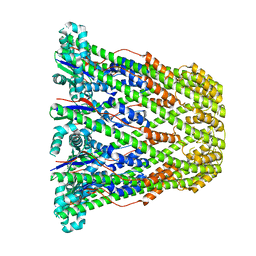 | |
5FZG
 
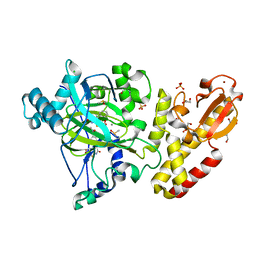 | | Crystal structure of the catalytic domain of human JARID1B in complex with MC3948 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Gileadi, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, Srikannathasan, V, von Delft, F, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Rotili, D, Mai, A, Oppermann, U. | | Deposit date: | 2016-03-14 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with Mc3948
To be Published
|
|
