6J3O
 
 | | Crystal structure of the human PCAF bromodomain in complex with compound 12 | | Descriptor: | 3-methyl-2-[[(3~{R})-1-methylpiperidin-3-yl]amino]-5~{H}-pyrrolo[3,2-d]pyrimidin-4-one, Histone acetyltransferase KAT2B | | Authors: | Huang, L.Y, Li, H, Li, L.L, Niu, L, Seupel, R, Wu, C.Y, Li, G.B, Yu, Y.M, Brennan, P.E, Yang, S.Y. | | Deposit date: | 2019-01-05 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Discovery of Pyrrolo[3,2- d]pyrimidin-4-one Derivatives as a New Class of Potent and Cell-Active Inhibitors of P300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 62, 2019
|
|
6J3P
 
 | | Crystal structure of the human GCN5 bromodomain in complex with compound (R,R)-36n | | Descriptor: | 2-{[(3R,5R)-5-(2,3-dihydro-1,4-benzodioxin-6-yl)-1-methylpiperidin-3-yl]amino}-3-methyl-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, Histone acetyltransferase KAT2A | | Authors: | Huang, L.Y, Li, H, Niu, L, Wu, C.Y, Yu, Y.M, Li, L.L, Yang, S.Y. | | Deposit date: | 2019-01-05 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Discovery of Pyrrolo[3,2- d]pyrimidin-4-one Derivatives as a New Class of Potent and Cell-Active Inhibitors of P300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 62, 2019
|
|
8FA3
 
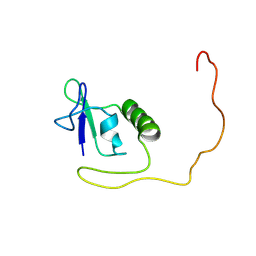 | |
3ZD2
 
 | | THE STRUCTURE OF THE TWO N-TERMINAL DOMAINS OF COMPLEMENT FACTOR H RELATED PROTEIN 1 SHOWS FORMATION OF A NOVEL DIMERISATION INTERFACE | | Descriptor: | 1,2-ETHANEDIOL, COMPLEMENT FACTOR H-RELATED PROTEIN 1 | | Authors: | Caesar, J.J.E, Goicoechea de Jorge, E, Pickering, M.C, Lea, S.M. | | Deposit date: | 2012-11-23 | | Release date: | 2013-03-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Dimerization of Complement Factor H-Related Proteins Modulates Complement Activation in Vivo.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3GT9
 
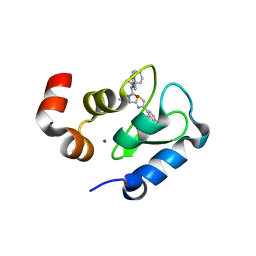 | | Structure of an ML-IAP/XIAP chimera bound to a peptidomimetic | | Descriptor: | Baculoviral IAP repeat-containing 7, N-{(1S)-1-cyclohexyl-2-[(2S)-2-(4-naphthalen-1-yl-1,3-thiazol-2-yl)pyrrolidin-1-yl]-2-oxoethyl}-N~2~-methyl-L-alaninamide, ZINC ION | | Authors: | Franklin, M.C, Fairbrother, W.J, Cohen, F. | | Deposit date: | 2009-03-27 | | Release date: | 2010-03-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Antagonists of inhibitor of apoptosis proteins based on thiazole amide isosteres.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3GTA
 
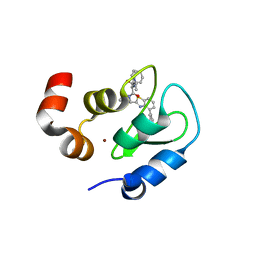 | | Structure of an ML-IAP/XIAP chimera bound to a peptidomimetic | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Baculoviral IAP repeat-containing 7, ... | | Authors: | Franklin, M.C, Fairbrother, W.J, Cohen, F. | | Deposit date: | 2009-03-27 | | Release date: | 2010-03-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Antagonists of inhibitor of apoptosis proteins based on thiazole amide isosteres.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
7DWC
 
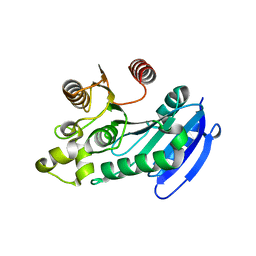 | | Bacteroides thetaiotaomicron VPI5482 BTAxe1 | | Descriptor: | Xylanase | | Authors: | Wang, L.Y, Wang, Y.L, Xin, F.J, Sun, L.C. | | Deposit date: | 2021-01-17 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Rational Design for Broadened Substrate Specificity and Enhanced Activity of a Novel Acetyl Xylan Esterase from Bacteroides thetaiotaomicron.
J.Agric.Food Chem., 69, 2021
|
|
6DHL
 
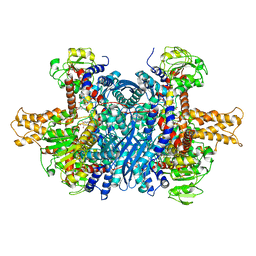 | | Bovine glutamate dehydrogenase complexed with epicatechin-3-gallate (ECG) | | Descriptor: | (2R,3S)-2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-3,4-dihydro-2H-chromen-3-yl 3,4,5-trihydroxybenzoate, Glutamate dehydrogenase 1, mitochondrial | | Authors: | Smith, T.J. | | Deposit date: | 2018-05-20 | | Release date: | 2018-07-25 | | Method: | X-RAY DIFFRACTION (3.624 Å) | | Cite: | Green tea polyphenols control dysregulated glutamate dehydrogenase in transgenic mice by hijacking the ADP activation site.
J. Biol. Chem., 286, 2011
|
|
8I16
 
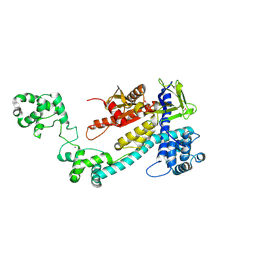 | | Crystal structure of the selenomethionine (SeMet)-derived Cas12g (D513A) mutant | | Descriptor: | Cas12g, ZINC ION | | Authors: | Zhang, B, Chen, J, Ye, Y.M, OuYang, S.Y. | | Deposit date: | 2023-01-12 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural transitions upon guide RNA binding and their importance in Cas12g-mediated RNA cleavage.
Plos Genet., 19, 2023
|
|
8I30
 
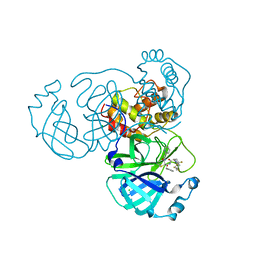 | | Crystal structure of the SARS-CoV-2 main protease in complex with 32j | | Descriptor: | (2~{R})-1-[4,4-bis(fluoranyl)cyclohexyl]carbonyl-4,4-bis(fluoranyl)-~{N}-[(2~{R},3~{S})-3-oxidanyl-4-oxidanylidene-1-phenyl-4-(pyridin-2-ylmethylamino)butan-2-yl]pyrrolidine-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION | | Authors: | Zeng, R, Huang, C, Xie, L.W, Wang, K, Liu, Y.Z, Yang, S.Y, Lei, J. | | Deposit date: | 2023-01-16 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and structure-activity relationship studies of novel alpha-ketoamide derivatives targeting the SARS-CoV-2 main protease.
Eur.J.Med.Chem., 259, 2023
|
|
8I3Q
 
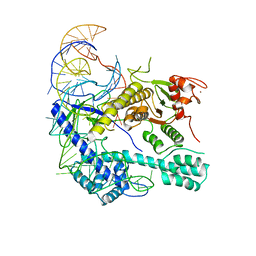 | |
1AMX
 
 | |
5H3D
 
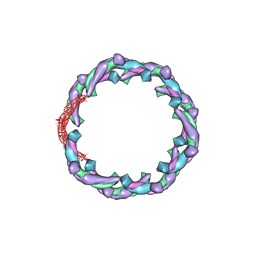 | | Helical structure of membrane tubules decorated by ACAP1 (BARPH doamin) protein by cryo-electron microscopy and MD simulation | | Descriptor: | Arf-GAP with coiled-coil, ANK repeat and PH domain-containing protein 1 | | Authors: | Chan, C, Pang, X.Y, Zhang, Y, Sun, F, Fan, J. | | Deposit date: | 2016-10-22 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | ACAP1 assembles into an unusual protein lattice for membrane deformation through multiple stages.
Plos Comput.Biol., 15, 2019
|
|
7E9B
 
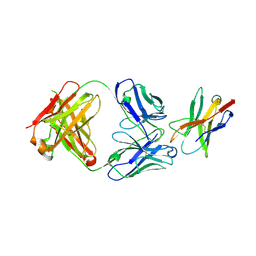 | |
1VYU
 
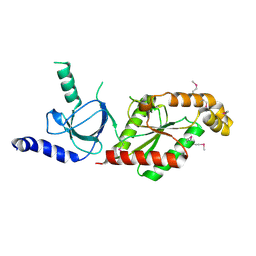 | | Beta3 subunit of Voltage-gated Ca2+-channel | | Descriptor: | CALCIUM CHANNEL BETA-3 SUBUNIT | | Authors: | Chen, Y.-H, Li, M.-H, Zhang, Y, He, L.-L, Yamada, Y, Fitzmaurice, A, Yang, S, Zhang, H, Liang, T, Yang, J. | | Deposit date: | 2004-05-07 | | Release date: | 2004-06-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of the Alpha(1)-Beta Subunit Interaction of Voltage-Gated Ca(2+) Channels
Nature, 429, 2004
|
|
7EXP
 
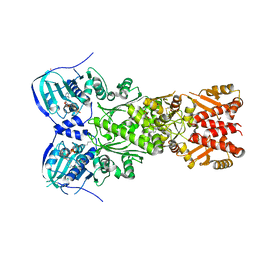 | | Crystal structure of zebrafish TRAP1 with AMPPNP and MitoQ | | Descriptor: | 2,3-dimethoxy-5-methyl-6-[10-(triphenyl-$l^{5}-phosphanyl)decyl]cyclohexa-2,5-diene-1,4-dione, COBALT (II) ION, MAGNESIUM ION, ... | | Authors: | Lee, H, Yoon, N.G, Kang, B.H, Lee, C. | | Deposit date: | 2021-05-28 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Mitoquinone Inactivates Mitochondrial Chaperone TRAP1 by Blocking the Client Binding Site.
J.Am.Chem.Soc., 143, 2021
|
|
1VYV
 
 | | beta4 subunit of Ca2+ channel | | Descriptor: | CALCIUM CHANNEL BETA-4SUBUNIT | | Authors: | Chen, Y.-H, Li, M.-H, Zhang, Y, He, L.-L, Yamada, Y, Fitzmaurice, A, Yang, S, Zhang, H, Liang, T, Yang, J. | | Deposit date: | 2004-05-07 | | Release date: | 2004-06-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of the Alpha(1)-Beta Subunit Interaction of Voltage-Gated Ca(2+) Channels
Nature, 429, 2004
|
|
1VYT
 
 | | beta3 subunit complexed with aid | | Descriptor: | CALCIUM CHANNEL BETA-3 SUBUNIT, VOLTAGE-DEPENDENT L-TYPE CALCIUM CHANNEL ALPHA-1C SUBUNIT | | Authors: | Chen, Y.-H, Li, M.-H, Zhang, Y, He, L.-L, Yamada, Y, Fitzmaurice, A, Yang, S, Zhang, H, Tong, L, Yang, J. | | Deposit date: | 2004-05-07 | | Release date: | 2004-06-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of the Alpha(1)-Beta Subunit Interaction of Voltage-Gated Ca(2+) Channels
Nature, 429, 2004
|
|
5XS2
 
 | |
5XQX
 
 | |
6YPE
 
 | |
7JU0
 
 | |
5J1D
 
 | | X-ray crystal structure of Phosphate binding protein (PBP) from Stenotrophomonas maltophilia | | Descriptor: | GLYCEROL, PHOSPHATE ION, Phosphate binding protein | | Authors: | Hatti, K, Gulati, A, Narayanswamy, S, Murthy, M.R.N. | | Deposit date: | 2016-03-29 | | Release date: | 2016-10-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Determination of crystal structures of proteins of unknown identity using a marathon molecular replacement procedure: structure of Stenotrophomonas maltophilia phosphate-binding protein.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5YZ9
 
 | | zinc finger domain of METTL3-METTL14 N6-methyladenosine methyltransferase | | Descriptor: | N6-adenosine-methyltransferase catalytic subunit, ZINC ION | | Authors: | Dong, X, Tang, C, Gong, Z, Yin, P, Huang, J.B. | | Deposit date: | 2017-12-13 | | Release date: | 2018-03-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Solution structure of the RNA recognition domain of METTL3-METTL14 N6-methyladenosine methyltransferase.
Protein Cell, 10, 2019
|
|
6N0M
 
 | |
