7X4I
 
 | |
7BTN
 
 | |
7VPN
 
 | |
7CMO
 
 | | Crystal structure of human inorganic pyrophosphatase | | Descriptor: | Inorganic pyrophosphatase | | Authors: | Hu, F, Huang, Z, Li, L. | | Deposit date: | 2020-07-28 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural and biochemical characterization of inorganic pyrophosphatase from Homo sapiens.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
3TK3
 
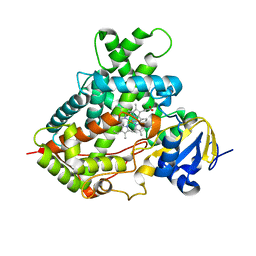 | | Cytochrome P450 2B4 mutant L437A in complex with 4-(4-chlorophenyl)imidazole | | Descriptor: | 4-(4-CHLOROPHENYL)IMIDAZOLE, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gay, S.C, Jang, H.H, Wilderman, P.R, Zhang, Q, Stout, C.D, Halpert, J.R. | | Deposit date: | 2011-08-25 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8001 Å) | | Cite: | Investigation by site-directed mutagenesis of the role of cytochrome P450 2B4 non-active-site residues in protein-ligand interactions based on crystal structures of the ligand-bound enzyme.
Febs J., 279, 2012
|
|
3U31
 
 | |
7Y86
 
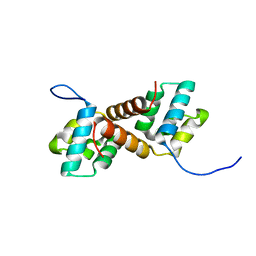 | | CcpS mutant | | Descriptor: | UPF0297 protein A7J08_00425 | | Authors: | Tang, J.S, Ran, T.T, Wang, W.W, Fan, H.J. | | Deposit date: | 2022-06-22 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A link between STK signalling and capsular polysaccharide synthesis in Streptococcus suis.
Nat Commun, 14, 2023
|
|
7Y8Z
 
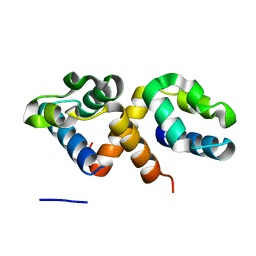 | | CcpS | | Descriptor: | UPF0297 protein A7J08_00425 | | Authors: | Tang, J.S, Ran, T.T, Wang, W.W, Fan, H.J. | | Deposit date: | 2022-06-24 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A link between STK signalling and capsular polysaccharide synthesis in Streptococcus suis.
Nat Commun, 14, 2023
|
|
3U3D
 
 | |
3R1B
 
 | | Open crystal structure of cytochrome P450 2B4 covalently bound to the mechanism-based inactivator tert-butylphenylacetylene | | Descriptor: | (4-tert-butylphenyl)acetaldehyde, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B4, ... | | Authors: | Gay, S.C, Zhang, H, Stout, C.D, Hollenberg, P.F, Halpert, J.R. | | Deposit date: | 2011-03-09 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Analysis of Mammalian Cytochrome P450 2B4 Covalently Bound to the Mechanism-Based Inactivator tert-Butylphenylacetylene: Insight into Partial Enzymatic Activity.
Biochemistry, 50, 2011
|
|
3RIG
 
 | |
3RIY
 
 | |
3R1A
 
 | | Closed crystal structure of cytochrome P450 2B4 covalently bound to the mechanism-based inactivator tert-butylphenylacetylene | | Descriptor: | (4-tert-butylphenyl)acetaldehyde, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gay, S.C, Zhang, H, Stout, C.D, Hollenberg, P.F, Halpert, J.R. | | Deposit date: | 2011-03-09 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Analysis of Mammalian Cytochrome P450 2B4 Covalently Bound to the Mechanism-Based Inactivator tert-Butylphenylacetylene: Insight into Partial Enzymatic Activity.
Biochemistry, 50, 2011
|
|
7VO8
 
 | | Structure of AtHPPD complexed with LSY-1 | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION, N-[6-[[9-(dimethylamino)-12H-benzo[a]phenoxazin-5-yl]amino]hexyl]-4-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-3-[oxidanyl(oxidanylidene)-$l^4-azanyl]benzamide | | Authors: | Yang, G.-F, Dong, J. | | Deposit date: | 2021-10-12 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | In vivo diagnostics of abiotic plant stress responses via in situ real-time fluorescence imaging
Plant Physiol., 190, 2022
|
|
8XN7
 
 | | Crystal structure of HPK1 kinase domain T165E,S171E phosphomimetic mutant in complex with compound 9f | | Descriptor: | 5-amino-2-((6-methoxy-2-methyl-1,2,3,4-tetrahydroisoquinolin-7-yl)amino)-8-(2-(trifluoromethyl)benzyl)pyrido[2,3-d]pyrimidin-7(8H)-one, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Huang, W.X, Liu, R, Ding, K. | | Deposit date: | 2023-12-29 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of 5-aminopyrido[2,3-d]pyrimidin-7(8H)-one derivatives as new hematopoietic progenitor kinase 1 (HPK1) inhibitors.
Eur.J.Med.Chem., 269, 2024
|
|
2QIC
 
 | | Crystal Structure of the ING1 PHD Finger in complex with a Histone H3K4ME3 peptide | | Descriptor: | H3K4ME3 PEPTIDE, Inhibitor of growth protein 1, ZINC ION | | Authors: | Pena, P.V, Champagne, K, Zhao, R, Kutateladze, T.G. | | Deposit date: | 2007-07-03 | | Release date: | 2008-05-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Histone H3K4me3 binding is required for the DNA repair and apoptotic activities of ING1 tumor suppressor.
J.Mol.Biol., 380, 2008
|
|
2RPN
 
 | | A crucial role for high intrinsic specificity in the function of yeast SH3 domains | | Descriptor: | Actin-binding protein, Actin-regulating kinase 1 | | Authors: | Stollar, E.J, Garcia, B, Chong, A, Forman-Kay, J, Davidson, A. | | Deposit date: | 2008-06-12 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural, functional, and bioinformatic studies demonstrate the crucial role of an extended peptide binding site for the SH3 domain of yeast Abp1p
J.Biol.Chem., 284, 2009
|
|
2HEN
 
 | |
3UAS
 
 | | Cytochrome P450 2B4 covalently bound to the mechanism-based inactivator 9-ethynylphenanthrene | | Descriptor: | 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Gay, S.C, Zhang, H, Shah, M.B, Stout, C.D, Halpert, J.R, Hollenberg, P.F. | | Deposit date: | 2011-10-21 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.939 Å) | | Cite: | Potent Mechanism-Based Inactivation of Cytochrome P450 2B4 by 9-Ethynylphenanthrene: Implications for Allosteric Modulation of Cytochrome P450 Catalysis.
Biochemistry, 52, 2013
|
|
2HWZ
 
 | | Fab fragment of Humanized anti-viral antibody MEDI-493 (Synagis TM) | | Descriptor: | Immunoglobulin Fab heavy chain, Immunoglobulin Fab light chain | | Authors: | Braden, B. | | Deposit date: | 2006-08-02 | | Release date: | 2007-08-14 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of a single tryptophan residue as critical for binding activity in a humanized monoclonal antibody against respiratory syncytial virus
To be Published
|
|
7EPX
 
 | | S protein of SARS-CoV-2 in complex with GW01 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Shen, Y.P, Zhang, Y.Y, Yan, R.H, Li, Y.N, Zhou, Q. | | Deposit date: | 2021-04-28 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Novel sarbecovirus bispecific neutralizing antibodies with exceptional breadth and potency against currently circulating SARS-CoV-2 variants and sarbecoviruses.
Cell Discov, 8, 2022
|
|
4F56
 
 | | The bicyclic intermediate structure provides insights into the desuccinylation mechanism of SIRT5 | | Descriptor: | 3-[(2R,3aR,5R,6R,6aR)-5-({[(S)-{[(S)-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}methyl)-2,6-dihydroxytetrahydrofuro[2,3-d][1,3]oxathiol-2-yl]propanoic acid, NAD-dependent lysine demalonylase and desuccinylase sirtuin-5, mitochondrial, ... | | Authors: | Zhou, Y, Hao, Q. | | Deposit date: | 2012-05-11 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Bicyclic Intermediate Structure Provides Insights into the Desuccinylation Mechanism of Human Sirtuin 5 (SIRT5)
J.Biol.Chem., 287, 2012
|
|
7WUI
 
 | | Tethered peptide activation mechanism of adhesion GPCRs ADGRG2 and ADGRG4 | | Descriptor: | Adhesion G-protein coupled receptor G2,mCherry, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Guo, S.C, He, Q.T, Xiao, P, Sun, J.P, Yu, X. | | Deposit date: | 2022-02-08 | | Release date: | 2022-04-27 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Tethered peptide activation mechanism of the adhesion GPCRs ADGRG2 and ADGRG4.
Nature, 604, 2022
|
|
7WUQ
 
 | | Tethered peptide activation mechanism of adhesion GPCRs ADGRG2 and ADGRG4 | | Descriptor: | Adhesion G-protein coupled receptor G2,mCherry, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Q.T, Guo, S.C, Xiao, P, Sun, J.P, Yu, X, Gou, L, Kong, L.L, Zhang, L. | | Deposit date: | 2022-02-09 | | Release date: | 2022-04-27 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Tethered peptide activation mechanism of the adhesion GPCRs ADGRG2 and ADGRG4.
Nature, 604, 2022
|
|
7WUJ
 
 | | Tethered peptide activation mechanism of adhesion GPCRs ADGRG2 and ADGRG4 | | Descriptor: | Adhesion G-protein coupled receptor G4,Uncharacterized protein, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Guo, S.C, Huang, S.M, He, Q.T, Xiao, P, Sun, J.P, Yu, X. | | Deposit date: | 2022-02-08 | | Release date: | 2022-04-27 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Tethered peptide activation mechanism of the adhesion GPCRs ADGRG2 and ADGRG4.
Nature, 604, 2022
|
|
