8U2Z
 
 | | TRPV1 in nanodisc bound with diC8-PIP2 in the dilated state | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, SODIUM ION, ... | | Authors: | Arnold, W.R, Julius, D, Cheng, Y. | | Deposit date: | 2023-09-06 | | Release date: | 2024-05-08 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of TRPV1 modulation by endogenous bioactive lipids.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8U30
 
 | | TRPV1 in nanodisc bound with diC8-PIP2 in the closed state | | Descriptor: | SODIUM ION, Transient receptor potential cation channel subfamily V member 1, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Arnold, W.R, Julius, D, Cheng, Y. | | Deposit date: | 2023-09-06 | | Release date: | 2024-05-08 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of TRPV1 modulation by endogenous bioactive lipids.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8U3L
 
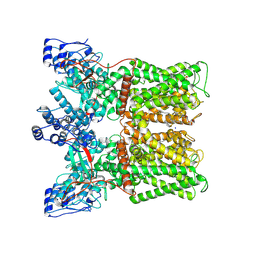 | | TRPV1 in nanodisc bound with empty vanilloid binding pocket at 25C | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, Transient receptor potential cation channel subfamily V member 1 | | Authors: | Arnold, W.R, Julius, D, Cheng, Y. | | Deposit date: | 2023-09-07 | | Release date: | 2024-05-08 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of TRPV1 modulation by endogenous bioactive lipids.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8U3J
 
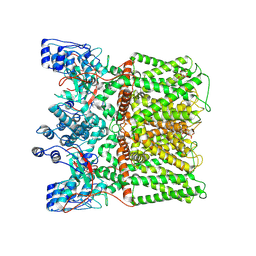 | | TRPV1 in nanodisc bound with empty vanilloid binding pocket at 4C | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, SODIUM ION, Transient receptor potential cation channel subfamily V member 1 | | Authors: | Arnold, W.R, Julius, D, Cheng, Y. | | Deposit date: | 2023-09-07 | | Release date: | 2024-05-08 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of TRPV1 modulation by endogenous bioactive lipids.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8U4D
 
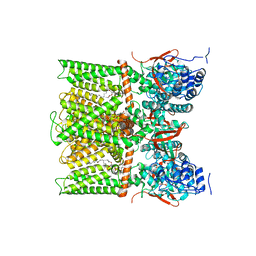 | | TRPV1 in nanodisc bound with PI-Br4, consensus structure | | Descriptor: | (2S)-2-[(9,10-dibromooctadecanoyl)oxy]-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9R,10S)-9,10-dibromooctadecanoate, SODIUM ION, Transient receptor potential cation channel subfamily V member 1 | | Authors: | Arnold, W.R, Julius, D, Cheng, Y. | | Deposit date: | 2023-09-10 | | Release date: | 2024-05-08 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structural basis of TRPV1 modulation by endogenous bioactive lipids.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8U3C
 
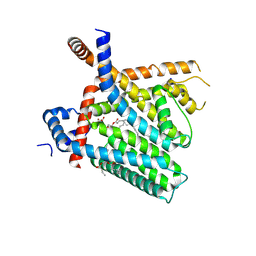 | | TRPV1 in nanodisc bound with PI-Br4 bound in Conformation 2 (monomer) | | Descriptor: | (2S)-2-[(9,10-dibromooctadecanoyl)oxy]-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9R,10S)-9,10-dibromooctadecanoate, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Arnold, W.R, Julius, D, Cheng, Y. | | Deposit date: | 2023-09-07 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural basis of TRPV1 modulation by endogenous bioactive lipids.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8U3A
 
 | | TRPV1 in nanodisc bound with PI-Br4 bound in Conformation 1 (monomer) | | Descriptor: | (2S)-2-[(9,10-dibromooctadecanoyl)oxy]-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9R,10S)-9,10-dibromooctadecanoate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, SODIUM ION, ... | | Authors: | Arnold, W.R, Julius, D, Cheng, Y. | | Deposit date: | 2023-09-07 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural basis of TRPV1 modulation by endogenous bioactive lipids.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8U43
 
 | | TRPV1 in nanodisc bound with PIP2-Br4 | | Descriptor: | (2S)-2-[(9,10-dibromooctadecanoyl)oxy]-3-{[(S)-hydroxy{[(1R,2R,3S,4R,5R,6S)-2,3,6-trihydroxy-4,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propyl (9R,10S)-9,10-dibromooctadecanoate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, SODIUM ION, ... | | Authors: | Arnold, W.R, Julius, D, Cheng, Y. | | Deposit date: | 2023-09-08 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis of TRPV1 modulation by endogenous bioactive lipids.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4AQB
 
 | | MBL-Ficolin Associated Protein-1, MAP-1 aka MAP44 | | Descriptor: | CALCIUM ION, MANNAN-BINDING LECTIN SERINE PROTEASE 1, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Skjoedt, M.O, Roversi, P, Hummelshoj, T, Palarasah, Y, Johnson, S, Lea, S.M, Garred, P. | | Deposit date: | 2012-04-16 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Crystal Structure and Functional Characterization of the Complement Regulator Mannose-Binding Lectin (Mbl)/Ficolin-Associated Protein-1 (Map-1).
J.Biol.Chem., 287, 2012
|
|
1FBR
 
 | |
2BH0
 
 | | Crystal structure of a SeMet derivative of EXPA from Bacillus subtilis at 2.5 angstrom | | Descriptor: | YOAJ | | Authors: | Petrella, S, Herman, R, Sauvage, E, Filee, P, Joris, B, Charlier, P. | | Deposit date: | 2005-01-06 | | Release date: | 2006-06-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure and Activity of Bacillus Subtilis Yoaj (Exlx1), a Bacterial Expansin that Promotes Root Colonization.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
1OHJ
 
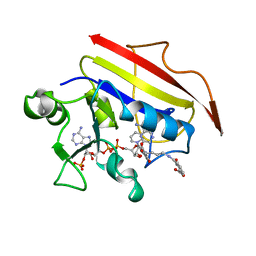 | | HUMAN DIHYDROFOLATE REDUCTASE, MONOCLINIC (P21) CRYSTAL FORM | | Descriptor: | DIHYDROFOLATE REDUCTASE, N-(4-CARBOXY-4-{4-[(2,4-DIAMINO-PTERIDIN-6-YLMETHYL)-AMINO]-BENZOYLAMINO}-BUTYL)-PHTHALAMIC ACID, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W. | | Deposit date: | 1997-09-17 | | Release date: | 1998-04-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Comparison of two independent crystal structures of human dihydrofolate reductase ternary complexes reduced with nicotinamide adenine dinucleotide phosphate and the very tight-binding inhibitor PT523.
Biochemistry, 36, 1997
|
|
4D86
 
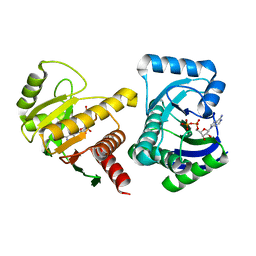 | | Human PARP14 (ARTD8, BAL2) - macro domains 1 and 2 in complex with adenosine-5-diphosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Poly [ADP-ribose] polymerase 14 | | Authors: | Karlberg, T, Thorsell, A.G, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Ekblad, T, Weigelt, J, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-01-10 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recognition of Mono-ADP-Ribosylated ARTD10 Substrates by ARTD8 Macrodomains.
Structure, 21, 2013
|
|
6UFI
 
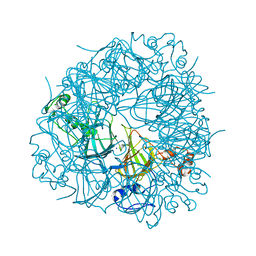 | | W96Y Oxalate Decarboxylase (Bacillus subtilis) | | Descriptor: | CHLORIDE ION, Cupin domain-containing protein, GLYCEROL, ... | | Authors: | Pastore, A.J, Burg, M.J, Twahir, U.T, Bruner, S.D, Angerhofer, A. | | Deposit date: | 2019-09-24 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Oxalate decarboxylase uses electron hole hopping for catalysis.
J.Biol.Chem., 297, 2021
|
|
7Z60
 
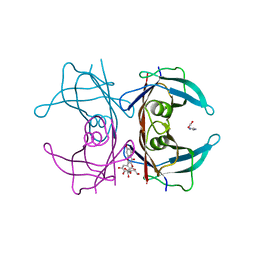 | | Human transthyretin (TTR) complexed with Quercetin 3-O-beta-D-galactoside | | Descriptor: | 1,2-ETHANEDIOL, 2-[3,4-bis(oxidanyl)phenyl]-3-[(2S,3R,4S,5R,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-5,7-bis(oxidanyl)chromen-4-one, IMIDAZOLE, ... | | Authors: | Ciccone, L, Braca, A, Orlandini, E, Nencetti, S. | | Deposit date: | 2022-03-10 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Antioxidant Quercetin 3-O-Glycosylated Plant Flavonols Contribute to Transthyretin Stabilization
Crystals, 12, 2022
|
|
8EGU
 
 | | Branched chain ketoacid dehydrogenase kinase complexes | | Descriptor: | (2~{S})-3-methyl-2-[pentanoyl-[[4-[2-(2~{H}-1,2,3,4-tetrazol-5-yl)phenyl]phenyl]methyl]amino]butanoic acid, ADENOSINE-5'-DIPHOSPHATE, SULFATE ION, ... | | Authors: | Liu, S. | | Deposit date: | 2022-09-13 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.923 Å) | | Cite: | Structural studies identify angiotensin II receptor blocker-like compounds as branched-chain ketoacid dehydrogenase kinase inhibitors.
J.Biol.Chem., 299, 2023
|
|
8EGD
 
 | | Branched chain ketoacid dehydrogenase kinase in complex with inhibitor | | Descriptor: | 5-(4-methoxyphenyl)-1H-tetrazole, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2022-09-12 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Structural studies identify angiotensin II receptor blocker-like compounds as branched-chain ketoacid dehydrogenase kinase inhibitors.
J.Biol.Chem., 299, 2023
|
|
8EGQ
 
 | | Branched chain ketoacid dehydrogenase kinase complexes | | Descriptor: | (2S)-2-ethyl-4-{[(2'M)-2'-(1H-tetrazol-5-yl)[1,1'-biphenyl]-4-yl]methyl}-3,4-dihydro-2H-pyrido[3,2-b][1,4]oxazine, ADENOSINE-5'-DIPHOSPHATE, SULFATE ION, ... | | Authors: | Liu, S. | | Deposit date: | 2022-09-13 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.955 Å) | | Cite: | Structural studies identify angiotensin II receptor blocker-like compounds as branched-chain ketoacid dehydrogenase kinase inhibitors.
J.Biol.Chem., 299, 2023
|
|
8EGF
 
 | | Branched chain ketoacid dehydrogenase kinase in complex with inhibitor | | Descriptor: | (5P)-5-(4'-methyl[1,1'-biphenyl]-2-yl)-1H-tetrazole, ADENOSINE-5'-DIPHOSPHATE, POTASSIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2022-09-12 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural studies identify angiotensin II receptor blocker-like compounds as branched-chain ketoacid dehydrogenase kinase inhibitors.
J.Biol.Chem., 299, 2023
|
|
8E8R
 
 | | 9H2 Fab-Sabin poliovirus 3 complex | | Descriptor: | 9H2 Fab heavy chain, 9H2 Fab light chain, Capsid protein VP1, ... | | Authors: | Charnesky, A.J. | | Deposit date: | 2022-08-25 | | Release date: | 2023-06-21 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | A human monoclonal antibody binds within the poliovirus receptor-binding site to neutralize all three serotypes.
Nat Commun, 14, 2023
|
|
8E8Y
 
 | | 9H2 Fab-Sabin poliovirus 2 complex | | Descriptor: | 9H2 Fab heavy chain, 9H2 Fab light chain, Capsid protein VP1, ... | | Authors: | Charnesky, A.J. | | Deposit date: | 2022-08-26 | | Release date: | 2023-06-21 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A human monoclonal antibody binds within the poliovirus receptor-binding site to neutralize all three serotypes.
Nat Commun, 14, 2023
|
|
8E8Z
 
 | | 9H2 Fab-Sabin poliovirus 1 complex | | Descriptor: | 9H2 Fab heavy chain, 9H2 Fab light chain, Capsid protein VP1, ... | | Authors: | Charnesky, A.J. | | Deposit date: | 2022-08-26 | | Release date: | 2023-06-21 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | A human monoclonal antibody binds within the poliovirus receptor-binding site to neutralize all three serotypes.
Nat Commun, 14, 2023
|
|
8E8X
 
 | | 9H2 Fab-Sabin poliovirus 3 complex | | Descriptor: | 9H2 Fab heavy chain, 9H2 Fab light chain, Capsid protein VP1, ... | | Authors: | Charnesky, A.J. | | Deposit date: | 2022-08-26 | | Release date: | 2023-06-21 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | A human monoclonal antibody binds within the poliovirus receptor-binding site to neutralize all three serotypes.
Nat Commun, 14, 2023
|
|
8E8S
 
 | | 9H2 Fab-poliovirus 2 complex | | Descriptor: | 9H2 Fab heavy chain, 9H2 Fab light chain, Capsid protein VP1, ... | | Authors: | Charnesky, A.J. | | Deposit date: | 2022-08-25 | | Release date: | 2023-06-21 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | A human monoclonal antibody binds within the poliovirus receptor-binding site to neutralize all three serotypes.
Nat Commun, 14, 2023
|
|
8E8L
 
 | | 9H2 Fab-poliovirus 1 complex | | Descriptor: | 9H2 Fab heavy chain, 9H2 Fab light chain, Capsid protein VP1, ... | | Authors: | Charnesky, A.J. | | Deposit date: | 2022-08-25 | | Release date: | 2023-06-21 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | A human monoclonal antibody binds within the poliovirus receptor-binding site to neutralize all three serotypes.
Nat Commun, 14, 2023
|
|
