1BWS
 
 | | CRYSTAL STRUCTURE OF GDP-4-KETO-6-DEOXY-D-MANNOSE EPIMERASE/REDUCTASE FROM ESCHERICHIA COLI A KEY ENZYME IN THE BIOSYNTHESIS OF GDP-L-FUCOSE | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTEIN (GDP-4-KETO-6-DEOXY-D-MANNOSE EPIMERASE/REDUCTASE) | | Authors: | Rizzi, M, Tonetti, M, Flora, A.D, Bolognesi, M. | | Deposit date: | 1998-09-25 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | GDP-4-keto-6-deoxy-D-mannose epimerase/reductase from Escherichia coli, a key enzyme in the biosynthesis of GDP-L-fucose, displays the structural characteristics of the RED protein homology superfamily.
Structure, 6, 1998
|
|
8SLU
 
 | | Crystal structure of human STEP (PTPN5) at cryogenic temperature (100 K) and high pressure (205 MPa) | | Descriptor: | GLYCEROL, SULFATE ION, Tyrosine-protein phosphatase non-receptor type 5 | | Authors: | Ebrahim, A, Guerrero, L, Riley, B.T, Kim, M, Huang, Q, Finke, A.D, Keedy, D.A. | | Deposit date: | 2023-04-24 | | Release date: | 2023-06-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Pushed to extremes: distinct effects of high temperature vs. pressure on the structure of an atypical phosphatase.
Biorxiv, 2023
|
|
8SLT
 
 | | Crystal structure of human STEP (PTPN5) at physiological temperature (310 K) and ambient pressure (0.1 MPa) | | Descriptor: | SULFATE ION, Tyrosine-protein phosphatase non-receptor type 5 | | Authors: | Ebrahim, A, Guerrero, L, Riley, B.T, Kim, M, Huang, Q, Finke, A.D, Keedy, D.A. | | Deposit date: | 2023-04-24 | | Release date: | 2023-06-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Pushed to extremes: distinct effects of high temperature vs. pressure on the structure of an atypical phosphatase.
Biorxiv, 2023
|
|
8SLS
 
 | | Crystal structure of human STEP (PTPN5) at cryogenic temperature (100 K) and ambient pressure (0.1 MPa) | | Descriptor: | GLYCEROL, SULFATE ION, Tyrosine-protein phosphatase non-receptor type 5 | | Authors: | Ebrahim, A, Guerrero, L, Riley, B.T, Kim, M, Huang, Q, Finke, A.D, Keedy, D.A. | | Deposit date: | 2023-04-24 | | Release date: | 2023-06-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Pushed to extremes: distinct effects of high temperature vs. pressure on the structure of an atypical phosphatase.
Biorxiv, 2023
|
|
8SZV
 
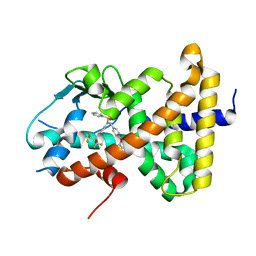 | | Crystal structure of pregnane X receptor ligand binding domain complexed with T0901317 analog SJPYT-318 | | Descriptor: | N-[4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)phenyl]-N-[(4-phenoxyphenyl)methyl]benzenesulfonamide, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Huber, A.D, Poudel, S, Miller, D.J, Li, Y, Chen, T. | | Deposit date: | 2023-05-30 | | Release date: | 2023-10-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ligand flexibility and binding pocket malleability cooperate to allow selective PXR activation by analogs of a promiscuous nuclear receptor ligand.
Structure, 31, 2023
|
|
8SPE
 
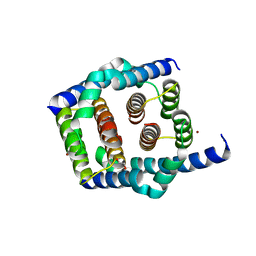 | | Crystal structure of Bax core domain BH3-groove dimer - tetrameric fraction P31 | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator BAX, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Miller, M.S, Cowan, A.D, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2023-05-03 | | Release date: | 2023-12-27 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Sequence differences between BAX and BAK core domains manifest as differences in their interactions with lipids.
Febs J., 291, 2024
|
|
8SPZ
 
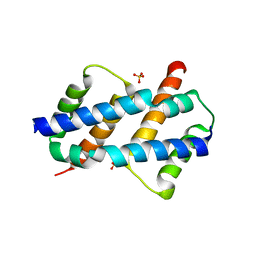 | | Crystal structure of Bax core domain BH3-groove dimer - hexameric fraction with dioctanoyl phosphatidylserine | | Descriptor: | Apoptosis regulator BAX, SULFATE ION | | Authors: | Cowan, A.D, Colman, P.M, Czabotar, P.E, Miller, M.S. | | Deposit date: | 2023-05-04 | | Release date: | 2023-12-27 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Sequence differences between BAX and BAK core domains manifest as differences in their interactions with lipids.
Febs J., 291, 2024
|
|
8SPF
 
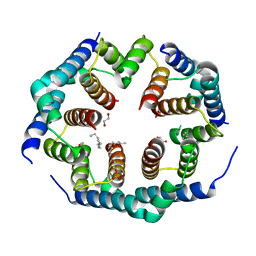 | | Crystal structure of Bax core domain BH3-groove dimer - hexameric fraction with 2-stearoyl lysoPC | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator BAX, DODECANE, ... | | Authors: | Cowan, A.D, Miller, M.S, Czabotar, P.E, Colman, P.M. | | Deposit date: | 2023-05-03 | | Release date: | 2023-12-27 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Sequence differences between BAX and BAK core domains manifest as differences in their interactions with lipids.
Febs J., 291, 2024
|
|
1D7K
 
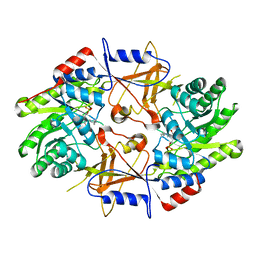 | | CRYSTAL STRUCTURE OF HUMAN ORNITHINE DECARBOXYLASE AT 2.1 ANGSTROMS RESOLUTION | | Descriptor: | HUMAN ORNITHINE DECARBOXYLASE | | Authors: | Almrud, J.J, Oliveira, M.A, Kern, A.D, Grishin, N.V, Phillips, M.A, Hackert, M.L. | | Deposit date: | 1999-10-18 | | Release date: | 2000-10-25 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human ornithine decarboxylase at 2.1 A resolution: structural insights to antizyme binding.
J.Mol.Biol., 295, 2000
|
|
8SVP
 
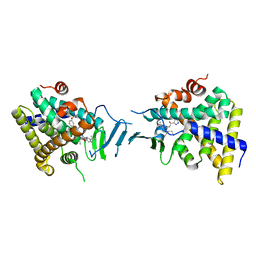 | | Crystal structure of pregnane X receptor ligand binding domain in complex with SJPYT-278 | | Descriptor: | (1P)-N-(5-tert-butyl-2-{[(3R)-hexan-3-yl]oxy}phenyl)-1-(2,5-dimethoxyphenyl)-5-methyl-1H-1,2,3-triazole-4-carboxamide, (1P)-N-(5-tert-butyl-2-{[(3S)-hexan-3-yl]oxy}phenyl)-1-(2,5-dimethoxyphenyl)-5-methyl-1H-1,2,3-triazole-4-carboxamide, Pregnane X receptor ligand binding domain fused to SRC-1 coactivator peptide | | Authors: | Garcia-Maldonado, E, Huber, A.D, Nithianantham, S, Miller, D.J, Chen, T. | | Deposit date: | 2023-05-17 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Chemical manipulation of an activation/inhibition switch in the nuclear receptor PXR.
Nat Commun, 15, 2024
|
|
8SVU
 
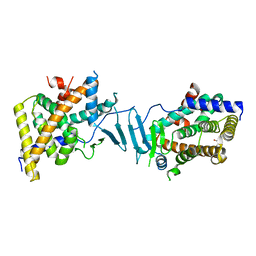 | | Crystal structure of the L428V mutant of pregnane X receptor ligand binding domain in apo form | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 1 group I member 2, Nuclear receptor coactivator 1 fusion protein,Nuclear receptor coactivator 1 | | Authors: | Garcia-Maldonado, E, Huber, A.D, Nithianantham, S, Miller, D.J, Chen, T. | | Deposit date: | 2023-05-17 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Chemical manipulation of an activation/inhibition switch in the nuclear receptor PXR.
Nat Commun, 15, 2024
|
|
8SVR
 
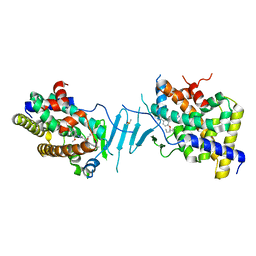 | | Crystal structure of pregnane X receptor ligand binding domain in complex with SJPYT-326 | | Descriptor: | (1P)-N-(5-tert-butyl-2-{[(3S)-hexan-3-yl]oxy}phenyl)-5-methyl-1-(2,4,5-trimethoxyphenyl)-1H-1,2,3-triazole-4-carboxamide, DIMETHYL SULFOXIDE, Pregnane X receptor ligand binding domain fused to SRC-1 coactivator peptide | | Authors: | Garcia-Maldonado, E, Huber, A.D, Nithianantham, S, Miller, D.J, Chen, T. | | Deposit date: | 2023-05-17 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Chemical manipulation of an activation/inhibition switch in the nuclear receptor PXR.
Nat Commun, 15, 2024
|
|
8SVS
 
 | | Crystal structure of pregnane X receptor ligand binding domain in complex with SJPYT-328 | | Descriptor: | (1P)-N-(5-tert-butyl-2-{[(3S)-hexan-3-yl]oxy}phenyl)-1-(2,4-dimethoxy-5-methylphenyl)-5-methyl-1H-1,2,3-triazole-4-carboxamide, Nuclear receptor subfamily 1 group I member 2, Nuclear receptor coactivator 1 fusion protein,Nuclear receptor coactivator 1 | | Authors: | Garcia-Maldonado, E, Huber, A.D, Nithianantham, S, Miller, D.J, Chen, T. | | Deposit date: | 2023-05-17 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Chemical manipulation of an activation/inhibition switch in the nuclear receptor PXR.
Nat Commun, 15, 2024
|
|
8SVT
 
 | | Crystal structure of pregnane X receptor ligand binding domain in complex with SJPYT-331 | | Descriptor: | DIMETHYL SULFOXIDE, Nuclear receptor subfamily 1 group I member 2, Nuclear receptor coactivator 1 fusion protein,Nuclear receptor coactivator 1, ... | | Authors: | Garcia-Maldonado, E, Huber, A.D, Nithianantham, S, Miller, D.J, Chen, T. | | Deposit date: | 2023-05-17 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Chemical manipulation of an activation/inhibition switch in the nuclear receptor PXR.
Nat Commun, 15, 2024
|
|
8SVQ
 
 | | Crystal structure of pregnane X receptor ligand binding domain in complex with SJPYT-312 | | Descriptor: | (1P)-N-(5-tert-butyl-2-{[(3S)-hexan-3-yl]oxy}phenyl)-1-(2,5-dimethoxyphenyl)-5-methyl-1H-1,2,3-triazole-4-carboxamide, DIMETHYL SULFOXIDE, Pregnane X receptor ligand binding domain fused to SRC-1 coactivator peptide | | Authors: | Garcia-Maldonado, E, Huber, A.D, Nithianantham, S, Miller, D.J, Chen, T. | | Deposit date: | 2023-05-17 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Chemical manipulation of an activation/inhibition switch in the nuclear receptor PXR.
Nat Commun, 15, 2024
|
|
8SVX
 
 | | Crystal structure of the L428V mutant of pregnane X receptor ligand binding domain in complex with SJPYT-331 | | Descriptor: | DIMETHYL SULFOXIDE, Nuclear receptor subfamily 1 group I member 2, Nuclear receptor coactivator 1 fusion protein,Nuclear receptor coactivator 1, ... | | Authors: | Garcia-Maldonado, E, Huber, A.D, Nithianantham, S, Miller, D.J, Chen, T. | | Deposit date: | 2023-05-17 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Chemical manipulation of an activation/inhibition switch in the nuclear receptor PXR.
Nat Commun, 15, 2024
|
|
8SVO
 
 | | Crystal structure of pregnane X receptor ligand binding domain in complex with SJPYT-310 | | Descriptor: | (1P)-N-(5-tert-butyl-2-{[(2S)-pentan-2-yl]oxy}phenyl)-1-(2-methoxy-5-methylphenyl)-5-methyl-1H-1,2,3-triazole-4-carboxamide, DIMETHYL SULFOXIDE, Pregnane X receptor ligand binding domain fused to SRC-1 coactivator peptide | | Authors: | Garcia-Maldonado, E, Huber, A.D, Nithianantham, S, Miller, D.J, Chen, T. | | Deposit date: | 2023-05-17 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Chemical manipulation of an activation/inhibition switch in the nuclear receptor PXR.
Nat Commun, 15, 2024
|
|
8SVN
 
 | | Crystal structure of the apo form of pregnane X receptor ligand binding domain | | Descriptor: | Pregnane X receptor ligand binding domain fused to SRC-1 coactivator peptide | | Authors: | Garcia-Maldonado, E, Huber, A.D, Nithianantham, S, Miller, D.J, Chen, T. | | Deposit date: | 2023-05-17 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chemical manipulation of an activation/inhibition switch in the nuclear receptor PXR.
Nat Commun, 15, 2024
|
|
1A87
 
 | | COLICIN N | | Descriptor: | COLICIN N | | Authors: | Vetter, I.R, Parker, M.W, Tucker, A.D, Lakey, J.H, Pattus, F, Tsernoglou, D. | | Deposit date: | 1998-04-03 | | Release date: | 1999-04-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of a colicin N fragment suggests a model for toxicity.
Structure, 6, 1998
|
|
1AXM
 
 | | HEPARIN-LINKED BIOLOGICALLY-ACTIVE DIMER OF FIBROBLAST GROWTH FACTOR | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ACIDIC FIBROBLAST GROWTH FACTOR | | Authors: | DiGabriele, A.D, Lax, I, Chen, D.I, Svahn, C.M, Jaye, M, Schlessinger, J, Hendrickson, W.A. | | Deposit date: | 1997-10-16 | | Release date: | 1998-04-22 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of a heparin-linked biologically active dimer of fibroblast growth factor.
Nature, 393, 1998
|
|
1CAQ
 
 | | X-RAY STRUCTURE OF HUMAN STROMELYSIN CATALYTIC DOMAIN COMPLEXES WITH NON-PEPTIDE INHIBITORS: IMPLICATION FOR INHIBITOR SELECTIVITY | | Descriptor: | 3-(1H-INDOL-3-YL)-2-[4-(4-PHENYL-PIPERIDIN-1-YL)-BENZENESULFONYLAMINO]-PROPIONIC ACID, CALCIUM ION, PROTEIN (STROMELYSIN-1), ... | | Authors: | Pavlovsky, A.G, Williams, M.G, Ye, Q.-Z, Ortwine, D.F, Purchase II, C.F, White, A.D, Dhanaraj, V, Roth, B.D, Johnson, L.L, Hupe, D, Humblet, C, Blundell, T.L. | | Deposit date: | 1999-02-23 | | Release date: | 1999-07-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structure of human stromelysin catalytic domain complexed with nonpeptide inhibitors: implications for inhibitor selectivity.
Protein Sci., 8, 1999
|
|
1BDN
 
 | |
1DJS
 
 | |
1DYW
 
 | | Biochemical and structural characterization of a divergent loop cyclophilin from Caenorhabditis elegans | | Descriptor: | CYCLOPHILIN 3 | | Authors: | Dornan, J, Page, A.P, Taylor, P, Wu, S.Y, Winter, A.D, Husi, H, Walkinshaw, M.D. | | Deposit date: | 2000-02-10 | | Release date: | 2000-06-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical and Structural Characterization of a Divergent Loop Cyclophilin from Caenorhabditis Elegans
J.Biol.Chem., 274, 1999
|
|
1B8Y
 
 | | X-RAY STRUCTURE OF HUMAN STROMELYSIN CATALYTIC DOMAIN COMPLEXED WITH NON-PEPTIDE INHIBITORS: IMPLICATIONS FOR INHIBITOR SELECTIVITY | | Descriptor: | CALCIUM ION, PROTEIN (STROMELYSIN-1), SULFATE ION, ... | | Authors: | Pavlovsky, A.G, Williams, M.G, Ye, Q.-Z, Ortwine, D.F, Purchase II, C.F, White, A.D, Dhanaraj, V, Roth, B.D, Johnson, L.L, Hupe, D, Humblet, C, Blundell, T.L. | | Deposit date: | 1999-02-03 | | Release date: | 1999-08-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of human stromelysin catalytic domain complexed with nonpeptide inhibitors: implications for inhibitor selectivity.
Protein Sci., 8, 1999
|
|
