7YPA
 
 | |
6L4R
 
 | | Crystal structure of Enterovirus D68 RdRp | | Descriptor: | RdRp | | Authors: | Wang, M.L, Li, L, Zhang, Y, Chen, Y.P, Su, D. | | Deposit date: | 2019-10-21 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Structure of the enterovirus D68 RNA-dependent RNA polymerase in complex with NADPH implicates an inhibitor binding site in the RNA template tunnel.
J.Struct.Biol., 211, 2020
|
|
4ZA1
 
 | | Crystal Structure of NosA Involved in Nosiheptide Biosynthesis | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, NosA | | Authors: | Liu, S, Guo, H, Zhang, T, Han, L, Yao, P, Zhang, Y, Rong, N, Yu, Y, Lan, W, Wang, C, Ding, J, Wang, R, Liu, W, Cao, C. | | Deposit date: | 2015-04-13 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based Mechanistic Insights into Terminal Amide Synthase in Nosiheptide-Represented Thiopeptides Biosynthesis
Sci Rep, 5, 2015
|
|
1ZM0
 
 | | Crystal Structure of the Carboxyl Terminal PH Domain of Pleckstrin To 2.1 Angstroms | | Descriptor: | Pleckstrin | | Authors: | Jackson, S.G, Zhang, Y, Zhang, K, Summerfield, R, Haslam, R.J, Junop, M.S. | | Deposit date: | 2005-05-09 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the carboxy-terminal PH domain of pleckstrin at 2.1 Angstroms.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
3L0C
 
 | |
3L0B
 
 | | Crystal structure of SCP1 phosphatase D206A mutant phosphoryl-intermediate | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, Carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1, MAGNESIUM ION | | Authors: | Zhang, M, Zhang, Y. | | Deposit date: | 2009-12-09 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and functional analysis of the phosphoryl transfer reaction mediated by the human small C-terminal domain phosphatase, Scp1.
Protein Sci., 19, 2010
|
|
3L0Y
 
 | | Crystal structure OF SCP1 phosphatase D98A mutant | | Descriptor: | Carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1, MAGNESIUM ION | | Authors: | Zhang, M, Zhang, Y. | | Deposit date: | 2009-12-10 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional analysis of the phosphoryl transfer reaction mediated by the human small C-terminal domain phosphatase, Scp1.
Protein Sci., 19, 2010
|
|
4MZ7
 
 | | Structural insight into dGTP-dependent activation of tetrameric SAMHD1 deoxynucleoside triphosphate triphosphohydrolase | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Zhu, C, Gao, W, Zhao, K, Qin, X, Zhang, Y, Peng, X, Zhang, L, Dong, Y, Zhang, W, Li, P, Wei, W, Gong, Y, Yu, X.F. | | Deposit date: | 2013-09-29 | | Release date: | 2013-11-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into dGTP-dependent activation of tetrameric SAMHD1 deoxynucleoside triphosphate triphosphohydrolase
Nat Commun, 4, 2013
|
|
7JVQ
 
 | | Cryo-EM structure of apomorphine-bound dopamine receptor 1 in complex with Gs protein | | Descriptor: | (6aR)-6-methyl-5,6,6a,7-tetrahydro-4H-dibenzo[de,g]quinoline-10,11-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Zhuang, Y, Xu, P, Mao, C, Wang, L, Krumm, B, Zhou, X.E, Huang, S, Liu, H, Cheng, X, Huang, X.-P, Sheng, D.-D, Xu, T, Liu, Y.-F, Wang, Y, Guo, J, Jiang, Y, Jiang, H, Melcher, K, Roth, B.L, Zhang, Y, Zhang, C, Xu, H.E. | | Deposit date: | 2020-08-22 | | Release date: | 2021-02-24 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the human D1 and D2 dopamine receptor signaling complexes.
Cell, 184, 2021
|
|
7JVP
 
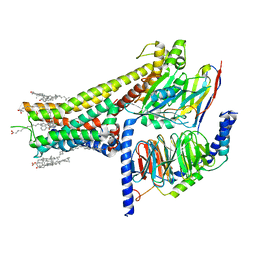 | | Cryo-EM structure of SKF-83959-bound dopamine receptor 1 in complex with Gs protein | | Descriptor: | (1R)-6-chloro-3-methyl-1-(3-methylphenyl)-2,3,4,5-tetrahydro-1H-3-benzazepine-7,8-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Zhuang, Y, Xu, P, Mao, C, Wang, L, Krumm, B, Zhou, X.E, Huang, S, Liu, H, Cheng, X, Huang, X.-P, Sheng, D.-D, Xu, T, Liu, Y.-F, Wang, Y, Guo, J, Jiang, Y, Jiang, H, Melcher, K, Roth, B.L, Zhang, Y, Zhang, C, Xu, H.E. | | Deposit date: | 2020-08-22 | | Release date: | 2021-02-24 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into the human D1 and D2 dopamine receptor signaling complexes.
Cell, 184, 2021
|
|
7JV5
 
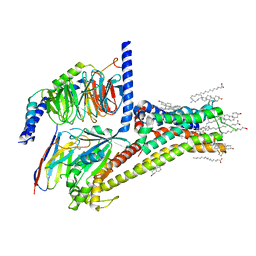 | | Cryo-EM structure of SKF-81297-bound dopamine receptor 1 in complex with Gs protein | | Descriptor: | (1R)-6-chloro-1-phenyl-2,3,4,5-tetrahydro-1H-3-benzazepine-7,8-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Zhuang, Y, Xu, P, Mao, C, Wang, L, Krumm, B, Zhou, X.E, Huang, S, Liu, H, Cheng, X, Huang, X.-P, Sheng, D.-D, Xu, T, Liu, Y.-F, Wang, Y, Guo, J, Jiang, Y, Jiang, H, Melcher, K, Roth, B.L, Zhang, Y, Zhang, C, Xu, H.E. | | Deposit date: | 2020-08-20 | | Release date: | 2021-02-24 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the human D1 and D2 dopamine receptor signaling complexes.
Cell, 184, 2021
|
|
7JVR
 
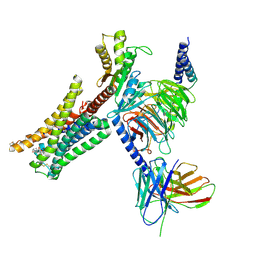 | | Cryo-EM structure of Bromocriptine-bound dopamine receptor 2 in complex with Gi protein | | Descriptor: | Antibody fragment ScFv16, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhuang, Y, Xu, P, Mao, C, Wang, L, Krumm, B, Zhou, X.E, Huang, S, Liu, H, Cheng, X, Huang, X.-P, Sheng, D.-D, Xu, T, Liu, Y.-F, Wang, Y, Guo, J, Jiang, Y, Jiang, H, Melcher, K, Roth, B.L, Zhang, Y, Zhang, C, Xu, H.E. | | Deposit date: | 2020-08-22 | | Release date: | 2021-02-24 | | Last modified: | 2021-03-31 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into the human D1 and D2 dopamine receptor signaling complexes.
Cell, 184, 2021
|
|
7CYQ
 
 | | Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Helicase, MAGNESIUM ION, ... | | Authors: | Yan, L, Ge, J, Zheng, L, Zhang, Y, Gao, Y, Wang, T, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-04 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-EM Structure of an Extended SARS-CoV-2 Replication and Transcription Complex Reveals an Intermediate State in Cap Synthesis.
Cell, 184, 2021
|
|
1P9Z
 
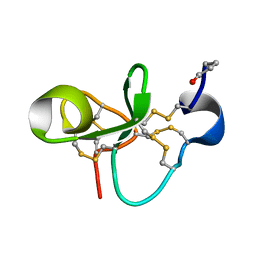 | | The Solution Structure of Antifungal Peptide Distinct With a Five-disulfide Motif from Eucommia ulmoides Oliver | | Descriptor: | Eucommia Antifungal peptide 2 | | Authors: | Huang, R.H, Xiang, Y, Tu, G.Z, Zhang, Y, Wang, D.C. | | Deposit date: | 2003-05-13 | | Release date: | 2004-05-25 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Eucommia Antifungal Peptide: A Novel Structural Model Distinct with a Five-Disulfide Motif.
Biochemistry, 43, 2004
|
|
6LH8
 
 | | Structure of aerolysin-like protein (Bombina maxima) | | Descriptor: | aerolysin-like protein | | Authors: | Bian, X.L, Wang, Q.Q, Li, X, Teng, M.Q, Zhang, Y. | | Deposit date: | 2019-12-07 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.729 Å) | | Cite: | A cellular endolysosome-modulating pore-forming protein from a toad is negatively regulated by its paralog under oxidizing conditions.
J.Biol.Chem., 295, 2020
|
|
6LHZ
 
 | | Structure of aerolysin-like protein (Bombina maxima) | | Descriptor: | aerolysin-like protein | | Authors: | Bian, X.L, Wang, Q.Q, Li, X, Teng, M.Q, Zhang, Y. | | Deposit date: | 2019-12-10 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | A cellular endolysosome-modulating pore-forming protein from a toad is negatively regulated by its paralog under oxidizing conditions.
J.Biol.Chem., 295, 2020
|
|
1GX0
 
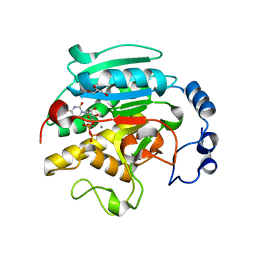 | | ALPHA-,1,3 GALACTOSYLTRANSFERASE - BETA-D-GALACTOSE COMPLEX | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | Authors: | Boix, E, Zhang, Y, Swaminathan, G.J, Brew, K, Acharya, K.R. | | Deposit date: | 2002-03-26 | | Release date: | 2003-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Ordered Binding of Donor and Acceptor Substrates to the Retaining Glycosyltransferase, Alpha -1,3 Galactosyltransferase
J.Biol.Chem., 277, 2002
|
|
1GWV
 
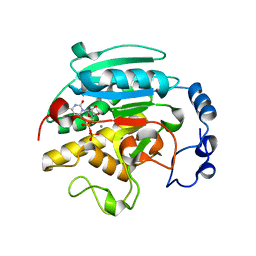 | | ALPHA-,1,3 GALACTOSYLTRANSFERASE - LACTOSE COMPLEX | | Descriptor: | MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Boix, E, Zhang, Y, Swaminathan, G.J, Brew, K, Acharya, K.R. | | Deposit date: | 2002-03-26 | | Release date: | 2003-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of Ordered Binding of Donor and Acceptor Substrates to the Retaining Glycosyltransferase, Alpha -1,3 Galactosyltransferase
J.Biol.Chem., 277, 2002
|
|
1GX4
 
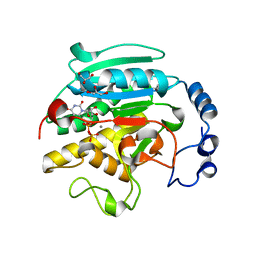 | | ALPHA-,1,3 GALACTOSYLTRANSFERASE - N-ACETYL LACTOSAMINE COMPLEX | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | Authors: | Boix, E, Zhang, Y, Swaminathan, G.J, Brew, K, Acharya, K.R. | | Deposit date: | 2002-03-27 | | Release date: | 2003-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural Basis of Ordered Binding of Donor and Acceptor Substrates to the Retaining Glycosyltransferase, Alpha -1,3 Galactosyltransferase
J.Biol.Chem., 277, 2002
|
|
1GWW
 
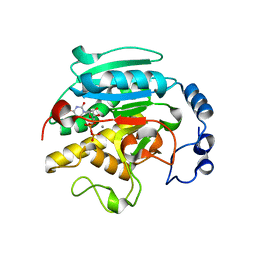 | | ALPHA-,1,3 GALACTOSYLTRANSFERASE - ALPHA-D-GLUCOSE COMPLEX | | Descriptor: | MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Boix, E, Zhang, Y, Swaminathan, G.J, Brew, K, Acharya, K.R. | | Deposit date: | 2002-03-26 | | Release date: | 2003-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Ordered Binding of Donor and Acceptor Substrates to the Retaining Glycosyltransferase, Alpha -1,3 Galactosyltransferase
J.Biol.Chem., 277, 2002
|
|
1IDS
 
 | | X-RAY STRUCTURE ANALYSIS OF THE IRON-DEPENDENT SUPEROXIDE DISMUTASE FROM MYCOBACTERIUM TUBERCULOSIS AT 2.0 ANGSTROMS RESOLUTIONS REVEALS NOVEL DIMER-DIMER INTERACTIONS | | Descriptor: | FE (III) ION, IRON SUPEROXIDE DISMUTASE | | Authors: | Cooper, J.B, Mcintyre, K, Wood, S.P, Zhang, Y, Young, D. | | Deposit date: | 1994-09-29 | | Release date: | 1994-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure analysis of the iron-dependent superoxide dismutase from Mycobacterium tuberculosis at 2.0 Angstroms resolution reveals novel dimer-dimer interactions.
J.Mol.Biol., 246, 1995
|
|
4KDO
 
 | | Crystal structure of the hemagglutinin of ferret-transmissible H5N1 virus in complex with human receptor analog LSTc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Lu, X, Shi, Y, Zhang, W, Zhang, Y, Qi, J, Gao, G.F. | | Deposit date: | 2013-04-25 | | Release date: | 2013-07-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Structure and receptor-binding properties of an airborne transmissible avian influenza A virus hemagglutinin H5 (VN1203mut).
Protein Cell, 4, 2013
|
|
4U86
 
 | | Human Pin1 with cysteine sulfonic acid 113 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Li, W, Zhang, Y. | | Deposit date: | 2014-08-01 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pin1 cysteine-113 oxidation inhibits its catalytic activity and cellular function in Alzheimer's disease.
Neurobiol.Dis., 76, 2015
|
|
3J3W
 
 | | Atomic model of the immature 50S subunit from Bacillus subtilis (state II-a) | | Descriptor: | 50S ribosomal protein L1, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | Authors: | Li, N, Guo, Q, Zhang, Y, Yuan, Y, Ma, C, Lei, J, Gao, N. | | Deposit date: | 2013-04-28 | | Release date: | 2013-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (10.7 Å) | | Cite: | Cryo-EM structures of the late-stage assembly intermediates of the bacterial 50S ribosomal subunit
Nucleic Acids Res., 41, 2013
|
|
4U84
 
 | | Human Pin1 with S-hydroxyl-cysteine 113 | | Descriptor: | POLYETHYLENE GLYCOL (N=34), Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Li, W, Zhang, Y. | | Deposit date: | 2014-08-01 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Pin1 cysteine-113 oxidation inhibits its catalytic activity and cellular function in Alzheimer's disease.
Neurobiol.Dis., 76, 2015
|
|
