7WRV
 
 | | The interface of JMB2002 Fab binds to SARS-CoV-2 Omicron Variant S | | Descriptor: | JMB2002 Fab heavy chain, JMB2002 Fab light chain, Spike glycoprotein | | Authors: | Yin, W, Xu, Y, Xu, P, Cao, X, Wu, C, Gu, C, He, X, Wang, X, Huang, S, Yuan, Q, Wu, K, Hu, W, Huang, Z, Liu, J, Wang, Z, Jia, F, Xia, K, Liu, P, Wang, X, Song, B, Zheng, J, Jiang, H, Cheng, X, Jiang, Y, Deng, S.J, Xu, H.E. | | Deposit date: | 2022-01-27 | | Release date: | 2022-03-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody.
Science, 375, 2022
|
|
1XT9
 
 | | Crystal Structure of Den1 in complex with Nedd8 | | Descriptor: | Neddylin, Sentrin-specific protease 8 | | Authors: | Reverter, D, Wu, K, Erdene, T.G, Pan, Z.Q, Wilkinson, K.D, Lima, C.D. | | Deposit date: | 2004-10-21 | | Release date: | 2004-12-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a Complex between Nedd8 and the Ulp/Senp Protease Family Member Den1.
J.Mol.Biol., 345, 2005
|
|
2L26
 
 | | Rv0899 from Mycobacterium tuberculosis contains two separated domains | | Descriptor: | Uncharacterized protein Rv0899/MT0922 | | Authors: | Shi, C, Li, J, Gao, Y, Wu, K, Huang, H, Tian, C. | | Deposit date: | 2010-08-12 | | Release date: | 2011-08-17 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structural Studies of Mycobacterium tuberculosis Rv0899 Reveal a Monomeric Membrane-Anchoring Protein with Two Separate Domains
J.Mol.Biol., 2011
|
|
8JR9
 
 | | Small molecule agonist (PCO371) bound to human parathyroid hormone receptor type 1 (PTH1R) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha-1, ... | | Authors: | Zhao, L, He, Q, Yuan, Q, Gu, Y, He, X, Shan, H, Li, J, Wang, K, Li, Y, Hu, W, Wu, K, Shen, J, Xu, H.E. | | Deposit date: | 2023-06-16 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Conserved class B GPCR activation by a biased intracellular agonist.
Nature, 621, 2023
|
|
1WD2
 
 | | Solution Structure of the C-terminal RING from a RING-IBR-RING (TRIAD) motif | | Descriptor: | Ariadne-1 protein homolog, ZINC ION | | Authors: | Capili, A.D, Edghill, E.L, Wu, K, Borden, K.L.B. | | Deposit date: | 2004-05-11 | | Release date: | 2004-07-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal RING Finger from a RING-IBR-RING/TRIAD Motif Reveals a Novel Zinc-binding Domain Distinct from a RING
J.Mol.Biol., 340, 2004
|
|
5Z10
 
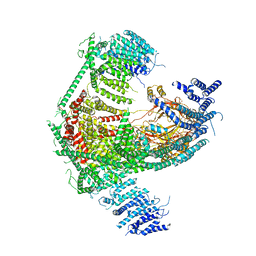 | | Structure of the mechanosensitive Piezo1 channel | | Descriptor: | Piezo-type mechanosensitive ion channel component 1 | | Authors: | Zhao, Q, Zhou, H, Chi, S, Wang, Y, Wang, J, Geng, J, Wu, K, Liu, W, Zhang, T, Dong, M.-Q, Wang, J, Li, X, Xiao, B. | | Deposit date: | 2017-12-22 | | Release date: | 2018-01-31 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Structure and mechanogating mechanism of the Piezo1 channel.
Nature, 554, 2018
|
|
6AGF
 
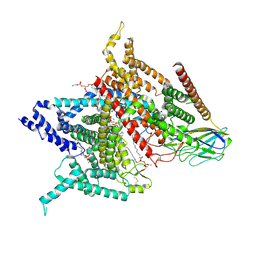 | | Structure of the human voltage-gated sodium channel Nav1.4 in complex with beta1 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pan, X.J, li, Z.Q, Zhou, Q, Shen, H.Z, Wu, K, Huang, X.S, Chen, J.F, Zhang, J.R, Zhu, X.C, Lei, J.L, Xiong, W, Gong, H.P, Xiao, B.L, Yan, N. | | Deposit date: | 2018-08-11 | | Release date: | 2018-10-10 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human voltage-gated sodium channel Nav1.4 in complex with beta 1.
Science, 362, 2018
|
|
8GY7
 
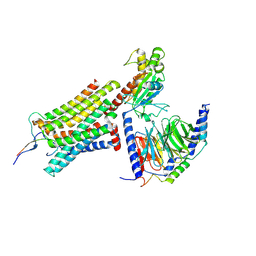 | | Cryo-EM structure of ACTH-bound melanocortin-2 receptor in complex with MRAP1 and Gs protein | | Descriptor: | CALCIUM ION, Corticotropin, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Luo, P, Feng, W.B, Ma, S.S, Dai, A.T, Yuan, Q.N, Wu, K, Yang, D.H, Wang, M.W, Xu, H.E, Jiang, Y. | | Deposit date: | 2022-09-21 | | Release date: | 2023-01-11 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of signaling regulation of the human melanocortin-2 receptor by MRAP1.
Cell Res., 33, 2023
|
|
4GG5
 
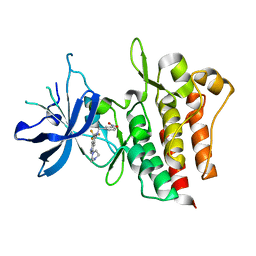 | | Crystal structure of CMET in complex with novel inhibitor | | Descriptor: | 3-(4-methylpiperazin-1-yl)-N-(3-nitrobenzyl)-7-(trifluoromethyl)quinolin-5-amine, Hepatocyte growth factor receptor | | Authors: | Liu, Q.F, Chen, T.T, Xu, Y.C. | | Deposit date: | 2012-08-05 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.423 Å) | | Cite: | Multisubstituted quinoxalines and pyrido[2,3-d]pyrimidines: Synthesis and SAR study as tyrosine kinase c-Met inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
8TQM
 
 | |
4GG7
 
 | | Crystal structure of cMET in complex with novel inhibitor | | Descriptor: | Hepatocyte growth factor receptor, N-(3-nitrobenzyl)-6-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]-2-(trifluoromethyl)pyrido[2,3-d]pyrimidin-4-amine | | Authors: | Liu, Q.F, Chen, T.T, Xu, Y.C. | | Deposit date: | 2012-08-06 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Multisubstituted quinoxalines and pyrido[2,3-d]pyrimidines: Synthesis and SAR study as tyrosine kinase c-Met inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
7UDK
 
 | | Crystal structure of designed helical repeat protein RPB_LRP2_R4 bound to LRPx4 peptide | | Descriptor: | 4xLRP, Designed helical repeat protein (DHR) RPB_LRP2_R4 | | Authors: | Chang, Y, Redler, R.L, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UDL
 
 | | Crystal structure of designed helical repeat protein RPB_PLP1_R6 bound to PLPx6 peptide | | Descriptor: | 1,2-ETHANEDIOL, 6xPLP Peptide, Designed helical repeat protein (DHR) RPB_PLP1_R6 | | Authors: | Chang, Y, Redler, R.L, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UDN
 
 | |
7UDM
 
 | | Crystal structure of designed helical repeat protein RPB_PLP1_R6 in alternative conformation 1 (with peptide) | | Descriptor: | 6xPLP, Designed helical repeat protein (DHR) RPB_PLP1_R6 | | Authors: | Chang, Y, Redler, R.L, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UDO
 
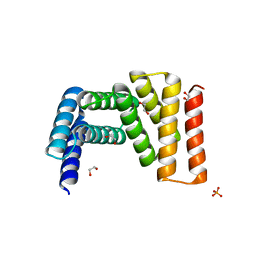 | | Crystal structure of designed helical repeat protein RPB_LRP2_R4 (proteolysis fragment?), forming pseudopolymeric filaments | | Descriptor: | 1,2-ETHANEDIOL, Designed helical repeat protein (DHR) RPB_LRP2_R4, PHOSPHATE ION | | Authors: | Redler, R.L, Chang, Y, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UDJ
 
 | | Crystal structure of designed helical repeat protein RPB_PEW3_R4 bound to PAWx4 peptide | | Descriptor: | 4xPAW peptide, De novo designed helical repeat protein RPB_PEW3_R4 | | Authors: | Redler, R.L, Chang, Y, Bhabha, G, Ekiert, D. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UE2
 
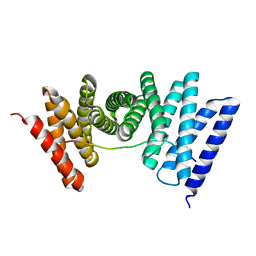 | |
9IYB
 
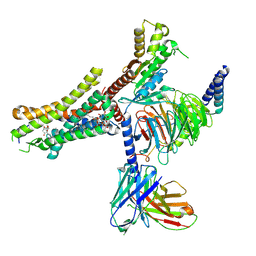 | | Cryo-EM Structure of the Prostaglandin D2 Receptor 2-PGD2 Coupled to G Protein | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Xu, J, Xu, Y, Wu, C, Xu, H.E. | | Deposit date: | 2024-07-30 | | Release date: | 2024-12-04 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Molecular basis of lipid and ligand regulation of prostaglandin receptor DP2.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7UHB
 
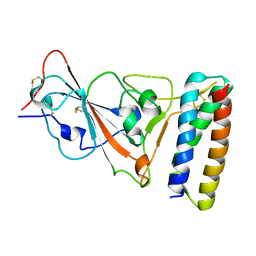 | |
7YW0
 
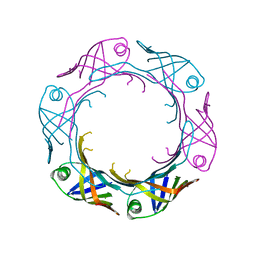 | | Bacteroides fragilis Hcp5 | | Descriptor: | Bacterodales T6SS protein TssD (Hcp) | | Authors: | Wen, Y, He, W, Bai, Y. | | Deposit date: | 2022-08-20 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure and assembly of type VI secretion system cargo delivery vehicle.
Cell Rep, 42, 2023
|
|
5C5S
 
 | |
6J8I
 
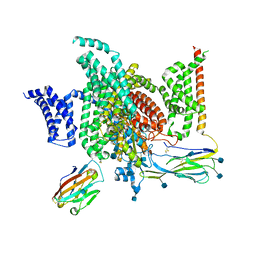 | | Structure of human voltage-gated sodium channel Nav1.7 in complex with auxiliary beta subunits, ProTx-II and tetrodotoxin (Y1755 up) | | Descriptor: | (1R,5R,6R,7R,9S,11S,12S,13S,14S)-3-amino-14-(hydroxymethyl)-8,10-dioxa-2,4-diazatetracyclo[7.3.1.1~7,11~.0~1,6~]tetradec-3-ene-5,9,12,13,14-pentol (non-preferred name), 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shen, H, Liu, D, Lei, J, Yan, N. | | Deposit date: | 2019-01-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of human Nav1.7 channel in complex with auxiliary subunits and animal toxins.
Science, 363, 2019
|
|
7F73
 
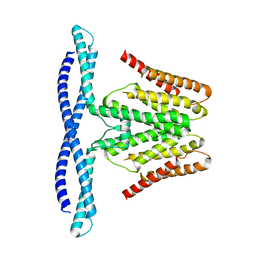 | | Cryo-EM structure of human TMEM120B | | Descriptor: | MCherry fluorescent protein,Transmembrane protein 120B | | Authors: | Ke, M, Wu, J, Yan, Z. | | Deposit date: | 2021-06-27 | | Release date: | 2021-09-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of human TMEM120A and TMEM120B.
Cell Discov, 7, 2021
|
|
8K88
 
 | | Structure of procaryotic Ago | | Descriptor: | DNA (41-mer), DNA/RNA (21-mer), MAGNESIUM ION, ... | | Authors: | Gao, X, Sun, D, Cui, S, Wang, Y. | | Deposit date: | 2023-07-29 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Nucleic acid-induced NADase activation of a short Sir2-associated prokaryotic Argonaute system.
Cell Rep, 43, 2024
|
|
