4GHQ
 
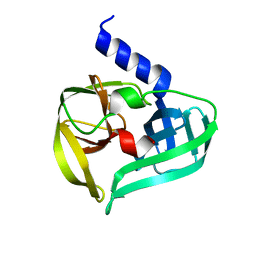 | | Crystal structure of EV71 3C proteinase | | Descriptor: | 3C proteinase | | Authors: | Chen, C, Wu, C, Cai, Q, Li, N, Peng, X, Cai, Y, Yin, K, Chen, X, Wang, X, Zhang, R, Liu, L, Chen, S, Li, J, Lin, T. | | Deposit date: | 2012-08-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of Enterovirus 71 3C proteinase (strain E2004104-TW-CDC) and its complex with rupintrivir
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7V66
 
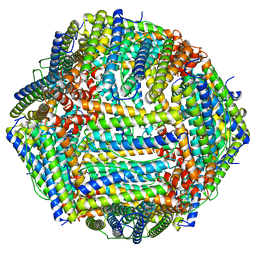 | | Structure of Apoferritin | | Descriptor: | Ferritin heavy chain | | Authors: | Zhang, X, Wu, C, Shi, H. | | Deposit date: | 2021-08-19 | | Release date: | 2022-10-05 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (1.89 Å) | | Cite: | Low-cooling-rate freezing in biomolecular cryo-electron microscopy for recovery of initial frames.
QRB Discov, 2, 2021
|
|
7DAE
 
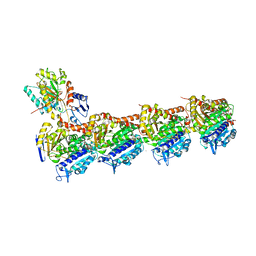 | | EPB in complex with tubulin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 7,11-DIHYDROXY-8,8,10,12,16-PENTAMETHYL-3-[1-METHYL-2-(2-METHYL-THIAZOL-4-YL)VINYL]-4,17-DIOXABICYCLO[14.1.0]HEPTADECANE-5,9-DIONE, CALCIUM ION, ... | | Authors: | Wu, C, Wang, Y. | | Deposit date: | 2020-10-16 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | High-resolution X-ray structure of three microtubule-stabilizing agents in complex with tubulin provide a rationale for drug design.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
7DAF
 
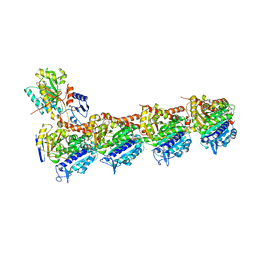 | | IXA in complex with tubulin | | Descriptor: | (1~{S},3~{S},7~{S},10~{R},11~{S},12~{S},16~{R})-8,8,10,12,16-pentamethyl-3-[(~{E})-1-(2-methyl-1,3-thiazol-4-yl)prop-1-en-2-yl]-7,11-bis(oxidanyl)-17-oxa-4-azabicyclo[14.1.0]heptadecane-5,9-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wu, C, Wang, Y. | | Deposit date: | 2020-10-16 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High-resolution X-ray structure of three microtubule-stabilizing agents in complex with tubulin provide a rationale for drug design.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
8ZW0
 
 | | Cryo-EM strcuture of Prostaglandin D2 Receptor DP1 activated by PGD2 | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, J, Xu, Y, Wu, C, Xu, H.E. | | Deposit date: | 2024-06-12 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Molecular basis for ligand recognition and receptor activation of the prostaglandin D2 receptor DP1.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8ZVZ
 
 | | Cryo-EM strcuture of Prostaglandin D2 Receptor DP1 activated by BW245C | | Descriptor: | 7-{(4S)-3-[(3R)-3-cyclohexyl-3-hydroxypropyl]-2,5-dioxoimidazolidin-4-yl}heptanoic acid, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, J, Xu, Y, Wu, C, Xu, H.E. | | Deposit date: | 2024-06-12 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Molecular basis for ligand recognition and receptor activation of the prostaglandin D2 receptor DP1.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
1U5S
 
 | | NMR structure of the complex between Nck-2 SH3 domain and PINCH-1 LIM4 domain | | Descriptor: | Cytoplasmic protein NCK2, PINCH protein, ZINC ION | | Authors: | Vaynberg, J, Fukuda, T, Vinogradova, O, Velyvis, A, Ng, L, Wu, C, Qin, J. | | Deposit date: | 2004-07-28 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of an ultraweak protein-protein complex and its crucial role in regulation of cell morphology and motility.
Mol.Cell, 17, 2005
|
|
9UWD
 
 | | Cryo-EM structure of inactive-DP1 | | Descriptor: | Prostaglandin D2 receptor,Soluble cytochrome b562 | | Authors: | Xu, J, Xu, Y, Wu, C, Xu, H.E. | | Deposit date: | 2025-05-12 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Molecular basis for ligand recognition and receptor activation of the prostaglandin D2 receptor DP1.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8ZEN
 
 | | Cryo-EM structure of Adr-2-Adbp-1-dsRBD0 complex | | Descriptor: | Adr-2-binding protein 1, Double-stranded RNA-specific adenosine deaminase adr-2, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Liu, Z.M, Mu, J.Q, Wu, C. | | Deposit date: | 2024-05-06 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Cryo-EM structure of Adr-2-Adbp-1-dsRBD0 complex
To Be Published
|
|
8ZEP
 
 | | Cryo-EM structure of Adr-2-Adbp-1-dsRBD2 complex | | Descriptor: | Adr-2-binding protein 1, Double-stranded RNA-specific adenosine deaminase adr-2, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Liu, Z.M, Mu, J.Q, Wu, C. | | Deposit date: | 2024-05-06 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cryo-EM structure of Adr-2-Adbp-1-dsRBD2 complex
To Be Published
|
|
8ZEO
 
 | | Cryo-EM structure of Adr-2-Adbp-1-dsRBD1 complex | | Descriptor: | Adr-2-binding protein 1, Double-stranded RNA-specific adenosine deaminase adr-2, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Liu, Z.M, Mu, J.Q, Wu, C. | | Deposit date: | 2024-05-06 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Cryo-EM structure of Adr-2-Adbp-1-dsRBD1 complex
To Be Published
|
|
9IWT
 
 | |
7FB8
 
 | | De Novo-Designed and Disulfide-Bridged Peptide Heterodimer - hd1 | | Descriptor: | ASP-ASP-LYS-ASP-CYS-ASP-GLU-TYR-CYS-LYS-LYS-THR-LYS-GLU-NH2, GLU-LE1-THR-GLY-HIS-ILE-GLU-GLY-PRO-THR-LE1-THR-LE1-HIS-CYS-LYS-NH2 | | Authors: | Yao, H, Yao, S, Zheng, Y, Moyer, A, Baker, D, Wu, C. | | Deposit date: | 2021-07-08 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | De novo design and directed folding of disulfide-bridged peptide heterodimers.
Nat Commun, 13, 2022
|
|
7FBA
 
 | | De Novo-Designed and Disulfide-Bridged Peptide Heterodimer - hd2 | | Descriptor: | ALA-LE1-CYS-GLU-CYS-GLY-PRO-THR-ARG-GLU-CYS-LYS-NH2, GLU-CYS-ARG-GLU-TYR-GLY-PRO-LE1-LYS-LE1-LE1-ALA-NH2 | | Authors: | Yao, H, Yao, S, Moyer, A, Zheng, Y, Baker, D, Wu, C. | | Deposit date: | 2021-07-08 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | De novo design and directed folding of disulfide-bridged peptide heterodimers.
Nat Commun, 13, 2022
|
|
8BQN
 
 | |
9IYB
 
 | | Cryo-EM Structure of the Prostaglandin D2 Receptor 2-PGD2 Coupled to G Protein | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Xu, J, Xu, Y, Wu, C, Xu, H.E. | | Deposit date: | 2024-07-30 | | Release date: | 2024-12-04 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Molecular basis of lipid and ligand regulation of prostaglandin receptor DP2.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8X7A
 
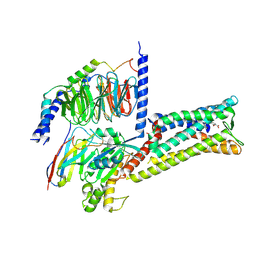 | | Treprostinil bound Prostacyclin Receptor G protein complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short,GNAS complex locus, ... | | Authors: | Wang, J.J, Jin, S, Zhang, H, Xu, Y, Hu, W, Jiang, Y, Chen, C, Wang, D.W, Xu, H.E, Wu, C. | | Deposit date: | 2023-11-23 | | Release date: | 2024-03-06 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Molecular recognition and activation of the prostacyclin receptor by anti-pulmonary arterial hypertension drugs.
Sci Adv, 10, 2024
|
|
8WSS
 
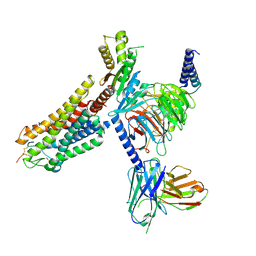 | | Cryo-EM structure of Melanin-Concentrating Hormone Receptor 1 with MCH | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhao, L, He, Q, Yuan, Q, Gu, Y, Shan, H, Hu, W, Wu, K, Xu, H.E, Zhang, Y, Wu, C. | | Deposit date: | 2023-10-17 | | Release date: | 2024-06-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Mechanisms of ligand recognition and activation of melanin-concentrating hormone receptors.
Cell Discov, 10, 2024
|
|
6YPZ
 
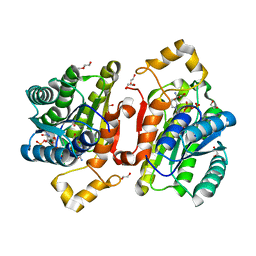 | | Promiscuous Reductase LugOII Catalyzes Keto-reduction at C1 during Lugdunomycin Biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Monooxygenase, ... | | Authors: | Xiao, X, Elsayed, S.S, Wu, C, van der Heul, H, Prota, A, Huang, J, Guo, R, Abrahams, J.P, van Wezel, G.P. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.
Acs Chem.Biol., 15, 2020
|
|
6YQ6
 
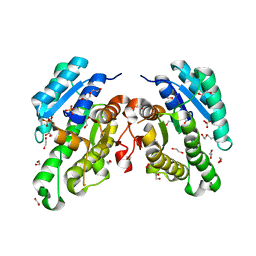 | | Promiscuous Reductase LugOII Catalyzes Keto-reduction at C1 during Lugdunomycin Biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Monooxygenase, ... | | Authors: | Xiao, X, Elsayed, S.S, Wu, C, van der Heul, H, Prota, A, Huang, J, Guo, R, Abrahams, J.P, van Wezel, G.P. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.
Acs Chem.Biol., 15, 2020
|
|
6YQ3
 
 | | Promiscuous Reductase LugOII Catalyzes Keto-reduction at C1 during Lugdunomycin Biosynthesis | | Descriptor: | (3~{R})-8-methoxy-3-methyl-3,6-bis(oxidanyl)-2,4-dihydrobenzo[a]anthracene-1,7,12-trione, 1,2-ETHANEDIOL, Monooxygenase, ... | | Authors: | Xiao, X, Elsayed, S.S, Wu, C, van der Heul, H, Prota, A, Huang, J, Guo, R, Abrahams, J.P, van Wezel, G.P. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.
Acs Chem.Biol., 15, 2020
|
|
6YQ0
 
 | | Promiscuous Reductase LugOII Catalyzes Keto-reduction at C1 during Lugdunomycin Biosynthesis | | Descriptor: | (3~{R})-8-methoxy-3-methyl-3-oxidanyl-2,4-dihydrobenzo[a]anthracene-1,7,12-trione, 1,2-ETHANEDIOL, Monooxygenase, ... | | Authors: | Xiao, X, Elsayed, S.S, Wu, C, van der Heul, H, Prota, A, Huang, J, Guo, R, Abrahams, J.P, van Wezel, G.P. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.
Acs Chem.Biol., 15, 2020
|
|
1HKS
 
 | | SOLUTION STRUCTURE OF THE DNA-BINDING DOMAIN OF DROSOPHILA HEAT SHOCK TRANSCRIPTION FACTOR | | Descriptor: | HEAT-SHOCK TRANSCRIPTION FACTOR | | Authors: | Vuister, G.W, Kim, S.-J, Orosz, A, Marquardt, J.L, Wu, C, Bax, A. | | Deposit date: | 1994-07-18 | | Release date: | 1994-09-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA-binding domain of Drosophila heat shock transcription factor.
Nat.Struct.Biol., 1, 1994
|
|
1HKT
 
 | | SOLUTION STRUCTURE OF THE DNA-BINDING DOMAIN OF DROSOPHILA HEAT SHOCK TRANSCRIPTION FACTOR | | Descriptor: | HEAT-SHOCK TRANSCRIPTION FACTOR | | Authors: | Vuister, G.W, Kim, S.-J, Orosz, A, Marquardt, J.L, Wu, C, Bax, A. | | Deposit date: | 1994-07-18 | | Release date: | 1994-09-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA-binding domain of Drosophila heat shock transcription factor.
Nat.Struct.Biol., 1, 1994
|
|
8X79
 
 | | MRE-269 bound Prostacyclin Receptor G protein complex | | Descriptor: | 2-[4-[(5,6-diphenylpyrazin-2-yl)-propan-2-yl-amino]butoxy]ethanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, J.J, Jin, S, Zhang, H, Xu, Y, Hu, W, Jiang, Y, Chen, C, Wang, D.W, Xu, H.E, Wu, C. | | Deposit date: | 2023-11-23 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Molecular recognition and activation of the prostacyclin receptor by anti-pulmonary arterial hypertension drugs.
Sci Adv, 10, 2024
|
|
