8R0J
 
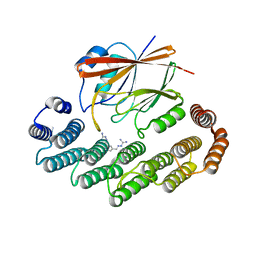 | | Crystal structure of the retromer complex VPS29/VPS35 with the ligand bis-1,3-phenyl guanylhydrazone, 2a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bis-1,3-phenyl guanylhydrazon, Vacuolar protein sorting-associated protein 29, ... | | Authors: | Milani, M, Fagnani, E. | | Deposit date: | 2023-10-31 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Stabilization of the retromer complex: Analysis of novel binding sites of bis-1,3-phenyl guanylhydrazone 2a to the VPS29/VPS35 interface.
Comput Struct Biotechnol J, 23, 2024
|
|
3UPF
 
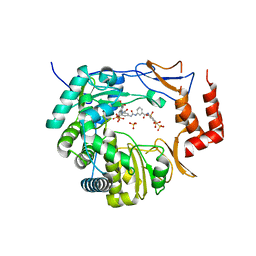 | | Crystal structure of murine norovirus RNA-dependent RNA polymerase bound to NF023 | | Descriptor: | 8-({3-[({3-[(4,6,8-trisulfonaphthalen-1-yl)carbamoyl]phenyl}carbamoyl)amino]benzoyl}amino)naphthalene-1,3,5-trisulfonic acid, RNA-dependent RNA polymerase, SULFATE ION | | Authors: | Milani, M, Mastrangelo, E, Bolognesi, M. | | Deposit date: | 2011-11-18 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Inhibition of Norovirus RNA-Dependent RNA Polymerases.
J.Mol.Biol., 419, 2012
|
|
3UQS
 
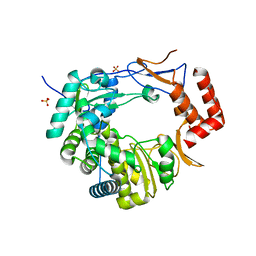 | |
3UR0
 
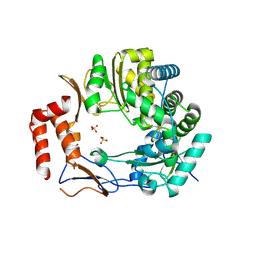 | | Crystal structures of murine norovirus RNA-dependent RNA polymerase in complex with Suramin | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFONIC ACID, RNA-dependent RNA polymerase, SULFATE ION | | Authors: | Milani, M, Mastrangelo, E, Bolognesi, M. | | Deposit date: | 2011-11-21 | | Release date: | 2012-05-02 | | Last modified: | 2012-05-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-Based Inhibition of Norovirus RNA-Dependent RNA Polymerases.
J.Mol.Biol., 419, 2012
|
|
4NRU
 
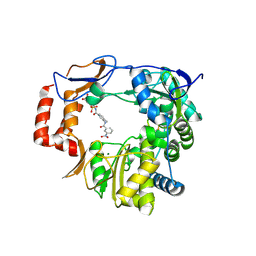 | | Murine Norovirus RNA-dependent-RNA-polymerase in complex with Compound 6, a suramin derivative | | Descriptor: | 4-({4-methyl-3-[(3-nitrobenzoyl)amino]benzoyl}amino)naphthalene-1,5-disulfonic acid, MAGNESIUM ION, RNA dependent RNA polymerase | | Authors: | Milani, M, Croci, R, Pezzullo, M, Tarantino, D, Mastrangelo, E, Bolognesi, M. | | Deposit date: | 2013-11-27 | | Release date: | 2014-10-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural bases of norovirus RNA dependent RNA polymerase inhibition by novel suramin-related compounds.
Plos One, 9, 2014
|
|
6G4H
 
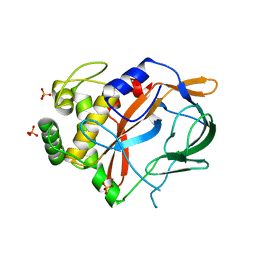 | | Crystal structure of the periplasmic domain of TgpA from Pseudomonas aeruginosa bound to ethylmercury | | Descriptor: | ETHYL MERCURY ION, PHOSPHATE ION, Protein-glutamine gamma-glutamyltransferase | | Authors: | Milani, M, Mastrangelo, E, Uruburu, M. | | Deposit date: | 2018-03-27 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional characterization of TgpA, a critical protein for the viability of Pseudomonas aeruginosa.
J.Struct.Biol., 205, 2019
|
|
6G49
 
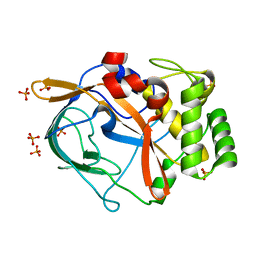 | | Crystal structure of the periplasmic domain of TgpA from Pseudomonas aeruginosa | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Protein-glutamine gamma-glutamyltransferase | | Authors: | Milani, M, Mastrangelo, E, Uruburu, M. | | Deposit date: | 2018-03-27 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional characterization of TgpA, a critical protein for the viability of Pseudomonas aeruginosa.
J.Struct.Biol., 205, 2019
|
|
4OXC
 
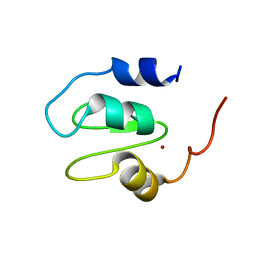 | | Crystal structure of XIAP BIR1 domain | | Descriptor: | E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Milani, M, Cossu, F, Mastrangelo, E. | | Deposit date: | 2014-02-05 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | NF023 binding to XIAP-BIR1: Searching drugs for regulation of the NF-kappa B pathway.
Proteins, 83, 2015
|
|
3CLX
 
 | | Crystal structure of XIAP BIR3 domain in complex with a Smac-mimetic compound, Smac005 | | Descriptor: | (3S,6S,7S,9aS)-6-{[(2S)-2-aminobutanoyl]amino}-N-(diphenylmethyl)-7-(hydroxymethyl)-5-oxooctahydro-1H-pyrrolo[1,2-a]azepine-3-carboxamide, Baculoviral IAP repeat-containing protein 4, ZINC ION | | Authors: | Milani, M, Mastrangelo, E, Cossu, F. | | Deposit date: | 2008-03-20 | | Release date: | 2008-10-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Targeting the X-linked inhibitor of apoptosis protein through 4-substituted azabicyclo[5.3.0]alkane smac mimetics. Structure, activity, and recognition principles.
J.Mol.Biol., 384, 2008
|
|
6QW3
 
 | | Calcium-bound gelsolin domain 2 | | Descriptor: | CALCIUM ION, Gelsolin | | Authors: | Scalone, E, Boni, F, Milani, M, Mastrangelo, E, de Rosa, M. | | Deposit date: | 2019-03-05 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-resolution crystal structure of gelsolin domain 2 in complex with the physiological calcium ion.
Biochem.Biophys.Res.Commun., 518, 2019
|
|
5FAE
 
 | | N184K pathological variant of gelsolin domain 2 (trigonal form) | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Boni, F, Milani, M, Ricagno, S, Bolognesi, M, de Rosa, M. | | Deposit date: | 2015-12-11 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of a novel renal amyloidosis due to N184K gelsolin variant.
Sci Rep, 6, 2016
|
|
3MUP
 
 | | cIAP1-BIR3 domain in complex with the Smac-mimetic compound Smac037 | | Descriptor: | (3S,6S,7R,9aS)-6-{[(2S)-2-aminobutanoyl]amino}-7-(2-aminoethyl)-N-(diphenylmethyl)-5-oxooctahydro-1H-pyrrolo[1,2-a]azepine-3-carboxamide, Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Cossu, F, Malvezzi, F, Canevari, G, Milani, M. | | Deposit date: | 2010-05-03 | | Release date: | 2010-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Recognition of Smac-mimetic compounds by the BIR domain of cIAP1
Protein Sci., 19, 2010
|
|
1H97
 
 | | Trematode hemoglobin from Paramphistomum epiclitum | | Descriptor: | Globin-3, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Pesce, A, Dewilde, S, Kiger, L, Milani, M, Ascenzi, P, Marden, M.C, Van Hauwaert, M.L, Vanfleteren, J, Moens, L, Bolognesi, M. | | Deposit date: | 2001-03-02 | | Release date: | 2001-06-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Very High Resolution Structure of a Trematode Hemoglobin Displaying a Tyrb10-Tyre7 Heme Distal Residue Pair and High Oxygen Affinity
J.Mol.Biol., 309, 2001
|
|
7NK0
 
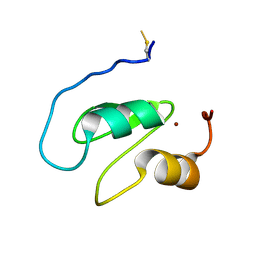 | | Structure of the BIR1 domain of cIAP2 | | Descriptor: | Baculoviral IAP repeat-containing protein 3, ZINC ION | | Authors: | Cossu, F, Milani, M, Mastrangelo, E, Mirdita, D. | | Deposit date: | 2021-02-17 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure-based identification of a new IAP-targeting compound that induces cancer cell death inducing NF-kappa B pathway.
Comput Struct Biotechnol J, 19, 2021
|
|
5FAF
 
 | | N184K pathological variant of gelsolin domain 2 (orthorhombic form) | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Boni, F, Milani, M, Ricagno, s, Bolognesi, M, de Rosa, M. | | Deposit date: | 2015-12-11 | | Release date: | 2016-10-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Molecular basis of a novel renal amyloidosis due to N184K gelsolin variant.
Sci Rep, 6, 2016
|
|
2WV9
 
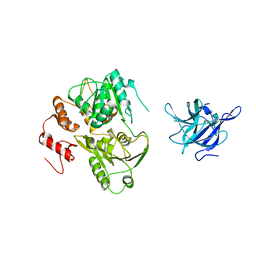 | | Crystal Structure of the NS3 protease-helicase from Murray Valley encephalitis virus | | Descriptor: | FLAVIVIRIN PROTEASE NS2B REGULATORY SUBUNIT, FLAVIVIRIN PROTEASE NS3 CATALYTIC SUBUNIT | | Authors: | Assenberg, R, Mastrangelo, E, Walter, T.S, Verma, A, Milani, M, Owens, R.J, Stuart, D.I, Grimes, J.M, Mancini, E.J. | | Deposit date: | 2009-10-15 | | Release date: | 2009-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of a Novel Conformational State of the Flavivirus Ns3 Protein: Implications for Polyprotein Processing and Viral Replication.
J.Virol., 83, 2009
|
|
5IQ6
 
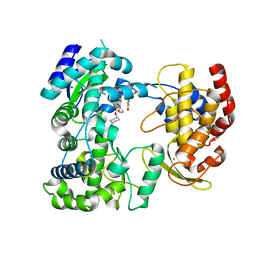 | | Crystal structure of Dengue virus serotype 3 RNA dependent RNA polymerase bound to HeE1-2Tyr, a new pyridobenzothizole inhibitor | | Descriptor: | N-[8-(cyclohexyloxy)-1-oxo-2-phenyl-1H-pyrido[2,1-b][1,3]benzothiazole-4-carbonyl]-L-tyrosine, RNA dependent RNA polymerase, ZINC ION | | Authors: | Tarantino, D, Mastrangelo, E, Milani, M. | | Deposit date: | 2016-03-10 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Targeting flavivirus RNA dependent RNA polymerase through a pyridobenzothiazole inhibitor.
Antiviral Res., 134, 2016
|
|
4EC4
 
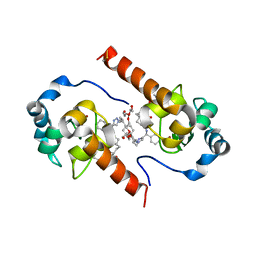 | | XIAP-BIR3 in complex with a potent divalent Smac mimetic | | Descriptor: | (3S,6S,7S,9aS,3'S,6'S,7'S,9a'S)-N,N'-(benzene-1,4-diylbis{butane-4,1-diyl-1H-1,2,3-triazole-1,4-diyl[(S)-phenylmethanediyl]})bis[7-(hydroxymethyl)-6-{[(2S)-2-(methylamino)butanoyl]amino}-5-oxooctahydro-1H-pyrrolo[1,2-a]azepine-3-carboxamide], 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Baculoviral IAP repeat-containing protein 4, ... | | Authors: | Mastrangelo, E, Cossu, F, Bolognesi, M, Milani, M. | | Deposit date: | 2012-03-26 | | Release date: | 2012-12-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural insight into inhibitor of apoptosis proteins recognition by a potent divalent smac-mimetic.
Plos One, 7, 2012
|
|
3OZ1
 
 | | cIAP1-BIR3 domain in complex with the Smac-mimetic compound Smac066 | | Descriptor: | (3S,6S,7R,9aS)-7-[2-(benzylamino)ethyl]-N-(diphenylmethyl)-6-{[(2S)-2-(methylamino)butanoyl]amino}-5-oxooctahydro-1H-pyrrolo[1,2-a]azepine-3-carboxamide, Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Cossu, F, Malvezzi, F, Mastrangelo, E, Canevari, G, Bolognesi, M, Milani, M. | | Deposit date: | 2010-09-24 | | Release date: | 2010-11-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Recognition of Smac-mimetic compounds by the BIR3 domain of cIAP1
Protein Sci., 2010
|
|
4EB9
 
 | | cIAP1-BIR3 in complex with a divalent Smac mimetic | | Descriptor: | (3S,6S,7S,9aS,3'S,6'S,7'S,9a'S)-N,N'-(benzene-1,4-diylbis{butane-4,1-diyl-1H-1,2,3-triazole-1,4-diyl[(S)-phenylmethanediyl]})bis[7-(hydroxymethyl)-6-{[(2S)-2-(methylamino)butanoyl]amino}-5-oxooctahydro-1H-pyrrolo[1,2-a]azepine-3-carboxamide], Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Cossu, F, Mastrangelo, E, Bolognesi, M, Milani, M. | | Deposit date: | 2012-03-23 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insight into inhibitor of apoptosis proteins recognition by a potent divalent smac-mimetic.
Plos One, 7, 2012
|
|
6Q9Z
 
 | | Crystal structure of the pathological G167R variant of calcium-free human gelsolin, | | Descriptor: | GLYCEROL, Gelsolin, SULFATE ION | | Authors: | Boni, F, Scalone, E, Milani, M, Eloise, M, de Rosa, M. | | Deposit date: | 2018-12-18 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The structure of N184K amyloidogenic variant of gelsolin highlights the role of the H-bond network for protein stability and aggregation properties.
Eur.Biophys.J., 49, 2020
|
|
6QBF
 
 | | Crystal structure of the pathological D187N variant of calcium-free human gelsolin. | | Descriptor: | GLYCEROL, Gelsolin, SODIUM ION, ... | | Authors: | Scalone, E, Boni, F, Milani, M, Eloise, M, de Rosa, M. | | Deposit date: | 2018-12-21 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.499 Å) | | Cite: | The structure of N184K amyloidogenic variant of gelsolin highlights the role of the H-bond network for protein stability and aggregation properties.
Eur.Biophys.J., 49, 2020
|
|
6Q9R
 
 | | Crystal structure of the pathological N184K variant of calcium-free human gelsolin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Scalone, E, Boni, F, Milani, M, Eloise, M, de Rosa, M. | | Deposit date: | 2018-12-18 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The structure of N184K amyloidogenic variant of gelsolin highlights the role of the H-bond network for protein stability and aggregation properties.
Eur.Biophys.J., 49, 2020
|
|
8QNS
 
 | |
2OXT
 
 | | Crystal structure of Meaban virus nucleoside-2'-O-methyltransferase | | Descriptor: | NUCLEOSIDE-2'-O-METHYLTRANSFERASE, S-ADENOSYLMETHIONINE | | Authors: | Mastrangelo, E, Milani, M, Bollati, M, Bolognesi, M. | | Deposit date: | 2007-02-21 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural bases for substrate recognition and activity in Meaban virus nucleoside-2'-O-methyltransferase
Protein Sci., 16, 2007
|
|
