2VSL
 
 | | Crystal Structure of XIAP BIR3 with a Bivalent Smac Mimetic | | Descriptor: | BACULOVIRAL IAP REPEAT-CONTAINING PROTEIN 4, PEPTIDE (MAA-LYS-PRO-PHE), POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2008-04-24 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Interaction of a Cyclic, Bivalent Smac Mimetic with the X-Linked Inhibitor of Apoptosis Protein.
Biochemistry, 47, 2008
|
|
6CJF
 
 | | Human dihydroorotate dehydrogenase bound to 4-quinoline carboxylic acid inhibitor 43 | | Descriptor: | 2-[4-(2-chloro-6-methylpyridin-3-yl)phenyl]-6-fluoro-3-methylquinoline-4-carboxylic acid, 3-[decyl(dimethyl)ammonio]propane-1-sulfonate, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2018-02-26 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 4-Quinoline Carboxylic Acids as Inhibitors of Dihydroorotate Dehydrogenase.
J. Med. Chem., 61, 2018
|
|
6DHS
 
 | | Structure of hnRNP H qRRM1,2 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein H | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2018-05-21 | | Release date: | 2018-09-12 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Differential Conformational Dynamics Encoded by the Inter-qRRM linker of hnRNP H.
J. Am. Chem. Soc., 2018
|
|
2X6P
 
 | | Crystal Structure of Coil Ser L19C | | Descriptor: | COIL SER L19C, ZINC ION | | Authors: | Chakraborty, S, Touw, D.S, Peacock, A.F.A, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2010-02-18 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Comparisons of Apo- and Metalated Three-Stranded Coiled Coils Clarify Metal Binding Determinants in Thiolate Containing Designed Peptides.
J.Am.Chem.Soc., 132, 2010
|
|
1YTS
 
 | | A LIGAND-INDUCED CONFORMATIONAL CHANGE IN THE YERSINIA PROTEIN TYROSINE PHOSPHATASE | | Descriptor: | SULFATE ION, YERSINIA PROTEIN TYROSINE PHOSPHATASE | | Authors: | Schubert, H.L, Stuckey, J.A, Fauman, E.B, Dixon, J.E, Saper, M.A. | | Deposit date: | 1995-04-07 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A ligand-induced conformational change in the Yersinia protein tyrosine phosphatase.
Protein Sci., 4, 1995
|
|
4I9O
 
 | | Crystal Structure of GACKIX L664C Tethered to 1-10 | | Descriptor: | 1,2-ETHANEDIOL, 1-{4-[4-chloro-3-(trifluoromethyl)phenyl]-4-hydroxypiperidin-1-yl}-3-sulfanylpropan-1-one, CREB-binding protein | | Authors: | Wang, N, Meagher, J.L, Stuckey, J.A, Mapp, A.K. | | Deposit date: | 2012-12-05 | | Release date: | 2013-03-06 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ordering a dynamic protein via a small-molecule stabilizer.
J.Am.Chem.Soc., 135, 2013
|
|
3PBJ
 
 | | Hydrolytic catalysis and structural stabilization in a designed metalloprotein | | Descriptor: | CHLORIDE ION, COIL SER L9L-Pen L23H, MERCURY (II) ION, ... | | Authors: | Zastrow, M.L, Peacock, A.F.A, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2010-10-20 | | Release date: | 2011-11-30 | | Last modified: | 2022-05-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Hydrolytic catalysis and structural stabilization in a designed metalloprotein.
Nat Chem, 4, 2012
|
|
2JGO
 
 | | Structure of the arsenated de novo designed peptide Coil Ser L9C | | Descriptor: | ARSENIC, COIL SER L9C, ZINC ION | | Authors: | Touw, D.S, Nordman, C.E, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2007-02-13 | | Release date: | 2007-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Identifying Important Structural Characteristics of Arsenic Resistance Proteins by Using Designed Three-Stranded Coiled Coils.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
4TW3
 
 | | Insights into Substrate and Metal Binding from the Crystal Structure of Cyanobacterial Aldehyde Deformylating Oxygenase with Substrate Bound | | Descriptor: | 1,2-ETHANEDIOL, Aldehyde decarbonylase, FE (III) ION, ... | | Authors: | Buer, B.C, Paul, B, Das, D, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2014-06-29 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into substrate and metal binding from the crystal structure of cyanobacterial aldehyde deformylating oxygenase with substrate bound.
Acs Chem.Biol., 9, 2014
|
|
2JK7
 
 | | XIAP BIR3 bound to a Smac Mimetic | | Descriptor: | (3S,6S,7Z,10AS)-N-(DIPHENYLMETHYL)-6-{[(2S)-2-(METHYLIDENEAMINO)BUTANOYL]AMINO}-5-OXO-1,2,3,5,6,9,10,10A-OCTAHYDROPYRROLO[1,2-A]AZOCINE-3-CARBOXAMIDE, BACULOVIRAL IAP REPEAT-CONTAINING PROTEIN 4, ZINC ION | | Authors: | Saito, N.G, Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2008-08-21 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structure-Based Design, Synthesis, Evaluation, and Crystallographic Studies of Conformationally Constrained Smac Mimetics as Inhibitors of the X-Linked Inhibitor of Apoptosis Protein (Xiap).
J.Med.Chem., 51, 2008
|
|
7TVA
 
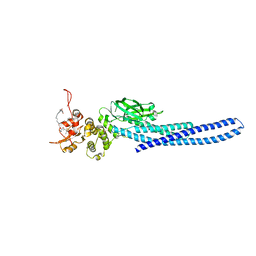 | | Stat5a Core in complex with AK2292 | | Descriptor: | DI(HYDROXYETHYL)ETHER, MALONATE ION, N-{5-[difluoro(phosphono)methyl]-1-benzothiophene-2-carbonyl}-3-methyl-L-valyl-L-prolyl-N-(5-{2-[(3R)-2,6-dioxopiperidin-3-yl]-1-oxo-2,3-dihydro-1H-isoindol-4-yl}pent-4-yn-1-yl)-N-methyl-N~3~-[4-(1,3-thiazol-2-yl)phenyl]-beta-alaninamide, ... | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2022-02-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.835 Å) | | Cite: | A selective small-molecule STAT5 PROTAC degrader capable of achieving tumor regression in vivo.
Nat.Chem.Biol., 19, 2023
|
|
7TVB
 
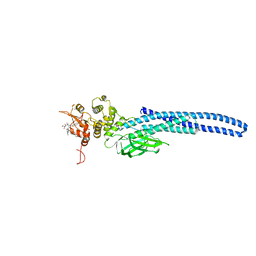 | | Stat5A Core in Complex with AK305 | | Descriptor: | GLYCEROL, N-{5-[difluoro(phosphono)methyl]-1-benzothiophene-2-carbonyl}-3-methyl-L-valyl-L-prolyl-N,N-dimethyl-N~3~-[4-(1,3-thiazol-2-yl)phenyl]-beta-alaninamide, Signal transducer and activator of transcription 5A | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2022-02-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | A selective small-molecule STAT5 PROTAC degrader capable of achieving tumor regression in vivo.
Nat.Chem.Biol., 19, 2023
|
|
7UC6
 
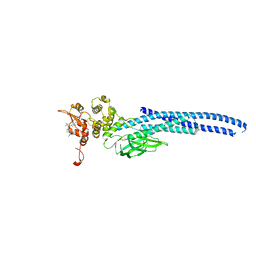 | | Stat5a Core in complex with Compound 12 | | Descriptor: | N-{5-[difluoro(phosphono)methyl]-1-benzothiophene-2-carbonyl}-3-methyl-L-valyl-L-prolyl-N~3~-(4-bromophenyl)-N,N-dimethyl-beta-alaninamide, Signal transducer and activator of transcription 5A | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2022-03-16 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Discovery of a Potent and Selective STAT5 PROTAC Degrader with Strong Antitumor Activity In Vivo in Acute Myeloid Leukemia.
J.Med.Chem., 66, 2023
|
|
7UBT
 
 | | Stat5a Core in complex with Compound 18 | | Descriptor: | GLYCEROL, N-{5-[difluoro(phosphono)methyl]-1-benzothiophene-2-carbonyl}-3-methyl-L-valyl-L-prolyl-N~3~-(1,3-benzothiazol-5-yl)-N,N-dimethyl-beta-alaninamide, Signal transducer and activator of transcription 5A | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2022-03-15 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | Discovery of a Potent and Selective STAT5 PROTAC Degrader with Strong Antitumor Activity In Vivo in Acute Myeloid Leukemia.
J.Med.Chem., 66, 2023
|
|
7UC7
 
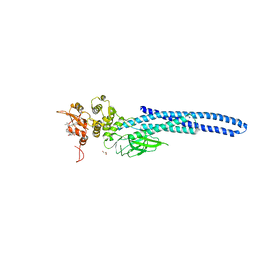 | | Stat5a Core in complex with Compound 17 | | Descriptor: | GLYCEROL, N-{5-[difluoro(phosphono)methyl]-1-benzothiophene-2-carbonyl}-3-methyl-L-valyl-L-prolyl-N~3~-(1-benzofuran-5-yl)-N,N-dimethyl-beta-alaninamide, Signal transducer and activator of transcription 5A | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2022-03-16 | | Release date: | 2023-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Discovery of a Potent and Selective STAT5 PROTAC Degrader with Strong Antitumor Activity In Vivo in Acute Myeloid Leukemia.
J.Med.Chem., 66, 2023
|
|
1XXP
 
 | | Yersinia YopH (residues 163-468) C403S binds phosphotyrosyl peptide at two sites | | Descriptor: | Hexapeptide ASP-ALA-ASP-GLU-PTR-CLE, Protein-tyrosine phosphatase yopH | | Authors: | Ivanov, M.I, Stuckey, J.A, Schubert, H.L, Saper, M.A, Bliska, J.B. | | Deposit date: | 2004-11-07 | | Release date: | 2005-03-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Two substrate-targeting sites in the Yersinia protein tyrosine phosphatase co-operate to promote bacterial virulence
Mol.Microbiol., 55, 2005
|
|
1XXV
 
 | | Yersinia YopH (residues 163-468) binds phosphonodifluoromethyl-Phe containing hexapeptide at two sites | | Descriptor: | Epidermal growth factor receptor derived peptide, Protein-tyrosine phosphatase yopH | | Authors: | Ivanov, M.I, Stuckey, J.A, Schubert, H.L, Saper, M.A, Bliska, J.B. | | Deposit date: | 2004-11-08 | | Release date: | 2005-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two substrate-targeting sites in the Yersinia protein tyrosine phosphatase co-operate to promote bacterial virulence
Mol.Microbiol., 55, 2005
|
|
6MCD
 
 | | Crystal Structure of tris-thiolate Pb(II) complex with adjacent water in a de novo Three Stranded Coiled Coil Peptide | | Descriptor: | LEAD (II) ION, Pb(II)(GRAND Coil Ser L12CL16A)-, ZINC ION | | Authors: | Tolbert, A.E, Ruckthong, L, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2018-08-31 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Heteromeric three-stranded coiled coils designed using a Pb(II)(Cys)3template mediated strategy.
Nat.Chem., 12, 2020
|
|
6MHD
 
 | | Glutathione S-Transferase Omega 1 bound to covalent inhibitor 44 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETONE, Glutathione S-transferase omega-1, ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure-Based Design of N-(5-Phenylthiazol-2-yl)acrylamides as Novel and Potent Glutathione S-Transferase Omega 1 Inhibitors.
J. Med. Chem., 62, 2019
|
|
6MHB
 
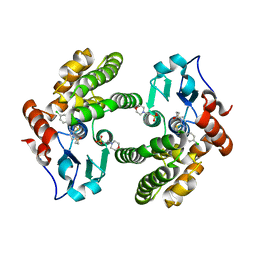 | | Glutathione S-Transferase Omega 1 bound to covalent inhibitor 18 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Glutathione S-transferase omega-1, N-[4-(4-chlorophenyl)-1,3-thiazol-2-yl]propanamide | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-Based Design of N-(5-Phenylthiazol-2-yl)acrylamides as Novel and Potent Glutathione S-Transferase Omega 1 Inhibitors.
J. Med. Chem., 62, 2019
|
|
6MHC
 
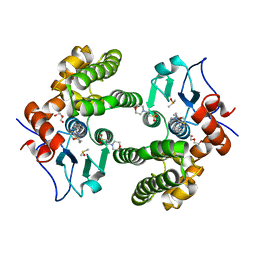 | | Glutathione S-Transferase Omega 1 bound to covalent inhibitor 37 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design of N-(5-Phenylthiazol-2-yl)acrylamides as Novel and Potent Glutathione S-Transferase Omega 1 Inhibitors.
J. Med. Chem., 62, 2019
|
|
6NJS
 
 | | Stat3 Core in complex with compound SD36 | | Descriptor: | Signal transducer and activator of transcription 3, [(2-{[(5S,8S,10aR)-3-acetyl-8-({(2S)-5-amino-1-[(diphenylmethyl)amino]-1,5-dioxopentan-2-yl}carbamoyl)-6-oxodecahydropyrrolo[1,2-a][1,5]diazocin-5-yl]carbamoyl}-1H-indol-5-yl)(difluoro)methyl]phosphonic acid (non-preferred name) | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2019-01-04 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Potent and Selective Small-Molecule Degrader of STAT3 Achieves Complete Tumor Regression In Vivo.
Cancer Cell, 36, 2019
|
|
4J09
 
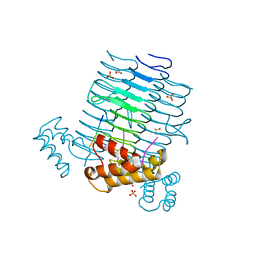 | | Crystal Structure of LpxA bound to RJPXD33 | | Descriptor: | 1,2-ETHANEDIOL, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, DIMETHYL SULFOXIDE, ... | | Authors: | Jenkins, R.J, Meagher, J.L, Stuckey, J.A, Dotson, G.D. | | Deposit date: | 2013-01-30 | | Release date: | 2014-04-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Recognition of Peptide RJPXD33 by Acyltransferases in Lipid A Biosynthesis.
J.Biol.Chem., 289, 2014
|
|
8GBN
 
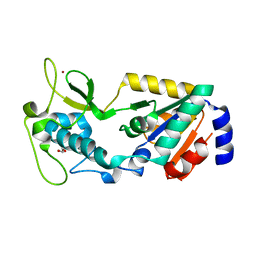 | | Structure of Apo Human SIRT5 P114T Mutant | | Descriptor: | 1,2-ETHANEDIOL, NAD-dependent protein deacylase sirtuin-5, mitochondrial, ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2023-02-26 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human SIRT5 variants with reduced stability and activity do not cause neuropathology in mice.
Iscience, 27, 2024
|
|
8GBL
 
 | | Structure of Apo Human SIRT5 | | Descriptor: | NAD-dependent protein deacylase sirtuin-5, mitochondrial, ZINC ION | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2023-02-26 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Human SIRT5 variants with reduced stability and activity do not cause neuropathology in mice.
Iscience, 27, 2024
|
|
