8H3E
 
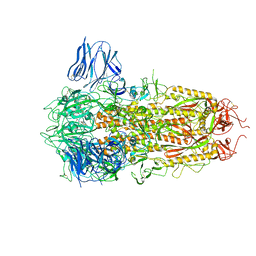 | | Complex structure of a small molecule (SPC-14) bound SARS-CoV-2 spike protein, closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-(6-nitro-2,3-dihydroindol-1-yl)-7-oxidanylidene-heptanoic acid, Spike glycoprotein,Fibritin | | Authors: | Meng, F, Wang, Q, Xie, Y, Ni, X, Huang, N. | | Deposit date: | 2022-10-08 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | In Silico Discovery of Small Molecule Modulators Targeting the Achilles' Heel of SARS-CoV-2 Spike Protein.
Acs Cent.Sci., 9, 2023
|
|
6AK4
 
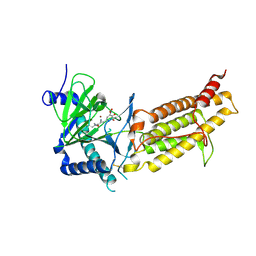 | | Crystal structure of human FTO in complex with small-molecule inhibitors | | Descriptor: | (~{E})-2-cyano-~{N},~{N}-diethyl-3-[3-nitro-4,5-bis(oxidanyl)phenyl]prop-2-enamide, Alpha-ketoglutarate-dependent dioxygenase FTO,Alpha-ketoglutarate-dependent dioxygenase FTO, ZINC ION | | Authors: | Wang, Y, Cao, R, Peng, S, Zhang, W, Huang, N. | | Deposit date: | 2018-08-30 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of entacapone as a chemical inhibitor of FTO mediating metabolic regulation through FOXO1.
Sci Transl Med, 11, 2019
|
|
6K0R
 
 | | Ruvbl1-Ruvbl2 with truncated domain II in complex with phosphorylated Cordycepin | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Zhang, W, Chen, L, Li, W, Ju, D, Huang, N, Zhang, E. | | Deposit date: | 2019-05-07 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Chemical perturbations reveal that RUVBL2 regulates the circadian phase in mammals.
Sci Transl Med, 12, 2020
|
|
8Y59
 
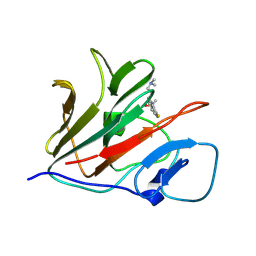 | | Crystal structure of TRIM21 PRYSPRY (D355A) in complex with (S)-hydroxyl-acepromazine. | | Descriptor: | (1~{S})-1-[10-[3-(dimethylamino)propyl]phenothiazin-2-yl]ethanol, E3 ubiquitin-protein ligase TRIM21 | | Authors: | Lu, P, Cheng, Y, Xue, L, Ren, X, Huang, N, Han, T. | | Deposit date: | 2024-01-31 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Selective degradation of multimeric proteins via chemically induced proximity to TRIM21.
To be published
|
|
8Y5B
 
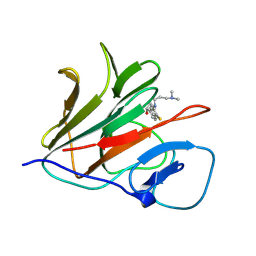 | | Crystal structure of TRIM21 PRYSPRY (D355A) in complex with (R)-hydroxyl-acepromazine. | | Descriptor: | (1~{R})-1-[10-[3-(dimethylamino)propyl]phenothiazin-2-yl]ethanol, E3 ubiquitin-protein ligase TRIM21 | | Authors: | Lu, P, Cheng, Y, Xue, L, Ren, X, Huang, N, Han, T. | | Deposit date: | 2024-01-31 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Selective degradation of multimeric proteins via chemically induced proximity to TRIM21.
To be published
|
|
8Y58
 
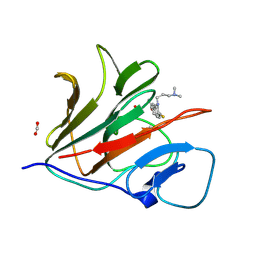 | | Crystal structure of TRIM21 PRYSPRY (D355A) in complex with acepromazine. | | Descriptor: | 1-[10-(3-DIMETHYLAMINO-PROPYL)-10H-PHENOTHIAZIN-2-YL]-ETHANONE, E3 ubiquitin-protein ligase TRIM21, FORMIC ACID | | Authors: | Lu, P, Cheng, Y, Xue, L, Ren, X, Huang, N, Han, T. | | Deposit date: | 2024-01-31 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Selective degradation of multimeric proteins via chemically induced proximity to TRIM21.
To be published
|
|
5KPP
 
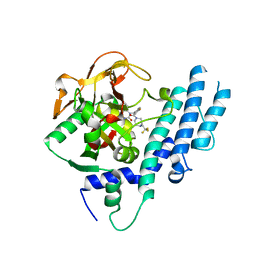 | | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor | | Descriptor: | 1-[[4-fluoranyl-3-[(3R)-3-methyl-4-[2,2,2-tris(fluoranyl)ethyl]piperazin-1-yl]carbonyl-phenyl]methyl]quinazoline-2,4-dione, Poly [ADP-ribose] polymerase 1 | | Authors: | Cao, R, Wang, Y.L, Zhou, J, Huang, N, Xu, B.L. | | Deposit date: | 2016-07-05 | | Release date: | 2016-11-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor
To Be Published
|
|
5KPO
 
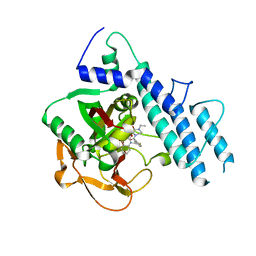 | | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor | | Descriptor: | 1-[[3-(4-ethyl-3-oxidanylidene-piperazin-1-yl)carbonyl-4-fluoranyl-phenyl]methyl]quinazoline-2,4-dione, Poly [ADP-ribose] polymerase 1 | | Authors: | Cao, R, Wang, Y.L, Zhou, J, Yao, H.P, Huang, N, Xu, B.L. | | Deposit date: | 2016-07-05 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor
To Be Published
|
|
5KPQ
 
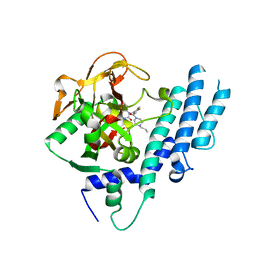 | | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor | | Descriptor: | 1-[[4-fluoranyl-3-[(3R)-3-methyl-4-propyl-piperazin-1-yl]carbonyl-phenyl]methyl]quinazoline-2,4-dione, Poly [ADP-ribose] polymerase 1 | | Authors: | Cao, R, Wang, Y.L, Zhou, J, Huang, N, Xu, B.L. | | Deposit date: | 2016-07-05 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of human PARP1 catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor
To Be Published
|
|
2Z8O
 
 | | Structural basis for the catalytic mechanism of phosphothreonine lyase | | Descriptor: | 27.5 kDa virulence protein, L(+)-TARTARIC ACID | | Authors: | Chen, L, Wang, H, Gu, L, Huang, N, Zhou, J.M, Chai, J. | | Deposit date: | 2007-09-07 | | Release date: | 2007-12-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the catalytic mechanism of phosphothreonine lyase.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2Z8P
 
 | | Structural basis for the catalytic mechanism of phosphothreonine lyase | | Descriptor: | (GLY)(GLU)(ALA)(TPO)(VAL)(PTR)(ALA), 27.5 kDa virulence protein | | Authors: | Chen, L, Wang, H, Gu, L, Huang, N, Zhou, J.M, Chai, J. | | Deposit date: | 2007-09-07 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the catalytic mechanism of phosphothreonine lyase.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2Z8N
 
 | | Structural basis for the catalytic mechanism of phosphothreonine lyase | | Descriptor: | 27.5 kDa virulence protein, SULFATE ION | | Authors: | Chen, L, Wang, H, Gu, L, Huang, N, Zhou, J.M, Chai, J. | | Deposit date: | 2007-09-07 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the catalytic mechanism of phosphothreonine lyase.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2Z8M
 
 | | Structural basis for the catalytic mechanism of phosphothreonine lyase | | Descriptor: | 27.5 kDa virulence protein | | Authors: | Chen, L, Wang, H, Gu, L, Huang, N, Zhou, J.M, Chai, J. | | Deposit date: | 2007-09-07 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the catalytic mechanism of phosphothreonine lyase.
Nat.Struct.Mol.Biol., 15, 2008
|
|
6KZI
 
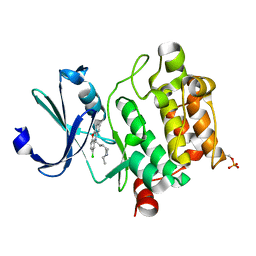 | | Crystal structure of Ser/Thr kinase Pim1 in complex with thioridazine derivatives | | Descriptor: | 4-(2-chloro-10H-phenoxazin-10-yl)-N,N-diethylbutan-1-amine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Wan, X, Li, W, Xie, Y, Huang, N. | | Deposit date: | 2019-09-24 | | Release date: | 2020-03-04 | | Last modified: | 2021-03-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
5C1Q
 
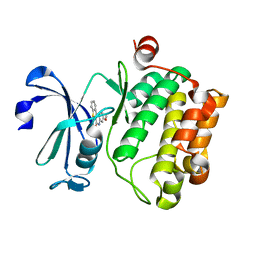 | | Serine/threonine-protein kinase pim-1 | | Descriptor: | 3-methoxy[1]benzothieno[2,3-c]quinolin-6(5H)-one, Serine/threonine-protein kinase pim-1 | | Authors: | Li, W, Wan, X, Huang, N. | | Deposit date: | 2015-06-15 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Serine/threonine-protein kinase pim-1
To Be Published
|
|
4RC3
 
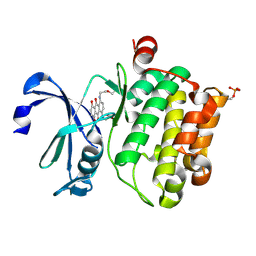 | |
4RC4
 
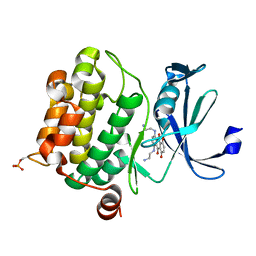 | |
4RBL
 
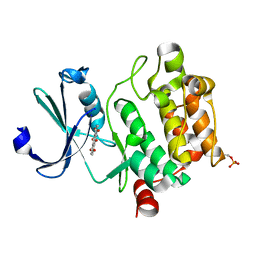 | | Crystal structure of Ser/Thr kinase Pim1 in complex with Mitoxantrone derivatives | | Descriptor: | 1,4,5,8-tetrahydroxyanthracene-9,10-dione, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Wan, X, Huang, N. | | Deposit date: | 2014-09-12 | | Release date: | 2015-03-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of Ser/Thr kinase Pim1 in complex with Mitoxantrone derivatives
To be Published, 2014
|
|
4RC2
 
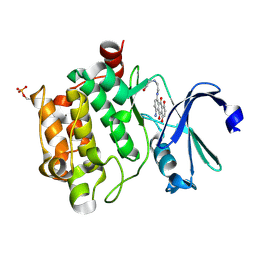 | |
5G06
 
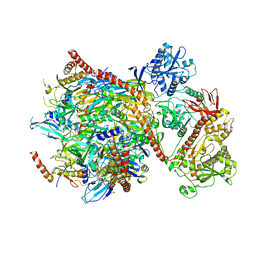 | | Cryo-EM structure of yeast cytoplasmic exosome | | Descriptor: | EXOSOME COMPLEX COMPONENT CSL4, EXOSOME COMPLEX COMPONENT MTR3, EXOSOME COMPLEX COMPONENT RRP4, ... | | Authors: | Liu, J.J, Niu, C.Y, Wu, Y, Tan, D, Wang, Y, Ye, M.D, Liu, Y, Zhao, W.W, Zhou, K, Liu, Q.S, Dai, J.B, Yang, X.R, Dong, M.Q, Huang, N, Wang, H.W. | | Deposit date: | 2016-03-17 | | Release date: | 2016-06-15 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryoem Structure of Yeast Cytoplasmic Exosome Complex.
Cell Res., 26, 2016
|
|
3HFJ
 
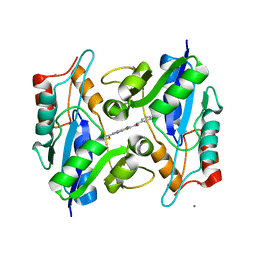 | | Bacillus anthracis nicotinate mononucleotide adenylytransferase (nadD) in complex with inhibitor CID 3289443 | | Descriptor: | 3-amino-N-(3-fluorophenyl)-6-thiophen-2-ylthieno[2,3-b]pyridine-2-carboxamide, CALCIUM ION, Nicotinate (Nicotinamide) nucleotide adenylyltransferase | | Authors: | Zhang, H, Huang, N, Eyobo, Y. | | Deposit date: | 2009-05-11 | | Release date: | 2009-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Targeting NAD biosynthesis in bacterial pathogens: Structure-based development of inhibitors of nicotinate mononucleotide adenylyltransferase NadD.
Chem.Biol., 16, 2009
|
|
5XYY
 
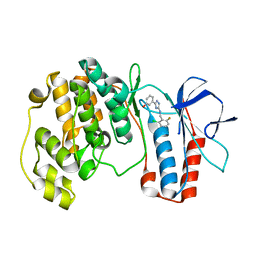 | | The structure of p38 alpha in complex with a triazol inhibitor | | Descriptor: | 3-(5-{[(2-chloro-6-fluorophenyl)methyl]amino}-4H-1,2,4-triazol-3-yl)phenol, Mitogen-activated protein kinase 14 | | Authors: | Wang, Y.L, Sun, Y.Z, Cao, R, Liu, D, Li, L, Qi, X.B, Huang, N. | | Deposit date: | 2017-07-11 | | Release date: | 2018-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In Silico Identification of a Novel Hinge-Binding Scaffold for Kinase Inhibitor Discovery.
J. Med. Chem., 60, 2017
|
|
6L15
 
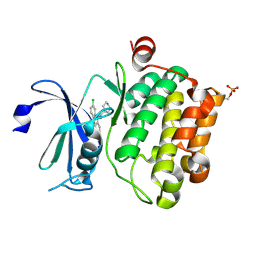 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 7-chloranyl-5-[3-[(3~{S})-piperidin-3-yl]propyl]pyrido[3,4-b][1,4]benzoxazine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6L12
 
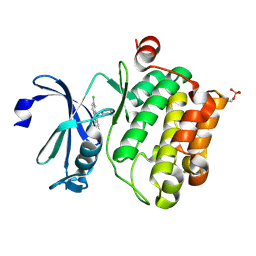 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 4-[(2-chloranylphenoxazin-10-yl)methyl]cyclohexan-1-amine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6L16
 
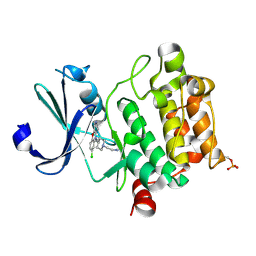 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 2-[4-[2-(7-chloranylpyrido[3,4-b][1,4]benzoxazin-5-yl)ethyl]piperidin-1-yl]ethanamine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
