5XU1
 
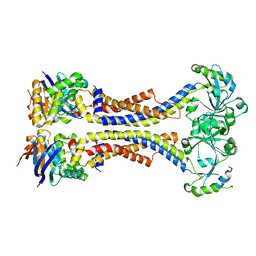 | | Structure of a non-canonical ABC transporter from Streptococcus pneumoniae R6 | | Descriptor: | ABC transporter ATP-binding protein, ABC transporter permeae, MAGNESIUM ION | | Authors: | Yang, H.B, Jiang, Y.L, Hou, W.T, Chen, M.T, Chen, Y, Zhou, C.Z. | | Deposit date: | 2017-06-22 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of a MacAB-like efflux pump from Streptococcus pneumoniae.
Nat Commun, 9, 2018
|
|
5XU0
 
 | | Structure of the membrane fusion protein Spr0693 from Streptococcus pneumoniae R6 | | Descriptor: | Membrane-fusion protein | | Authors: | Yang, H.B, Jiang, Y.L, Hou, W.T, Chen, M.T, Chen, Y, Zhou, C.Z. | | Deposit date: | 2017-06-22 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of a MacAB-like efflux pump from Streptococcus pneumoniae.
Nat Commun, 9, 2018
|
|
5Y2V
 
 | | Strcutrue of the full-length CcmR complexed with 2-OG from Synechocystis PCC6803 | | Descriptor: | 2-OXOGLUTARIC ACID, PHOSPHATE ION, Rubisco operon transcriptional regulator | | Authors: | Jiang, Y.L, Wang, X.P, Sun, H, Cheng, W, Han, S.J, Li, W.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2017-07-27 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Coordinating carbon and nitrogen metabolic signaling through the cyanobacterial global repressor NdhR.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5Y2W
 
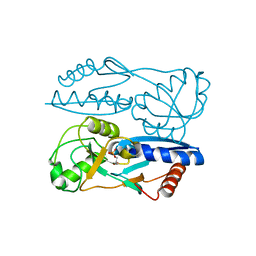 | | Structure of Synechocystis PCC6803 CcmR regulatory domain in complex with 2-PG | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Rubisco operon transcriptional regulator | | Authors: | Jiang, Y.L, Wang, X.P, Sun, H, Cheng, W, Cao, D.D, Han, S.J, Li, W.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2017-07-27 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Coordinating carbon and nitrogen metabolic signaling through the cyanobacterial global repressor NdhR.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7KDP
 
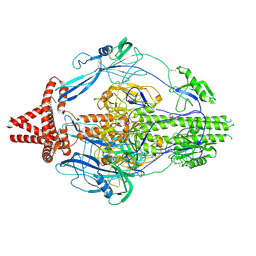 | | HCMV prefusion gB in complex with fusion inhibitor WAY-174865 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Envelope glycoprotein B, ... | | Authors: | Liu, Y, Heim, P.K, Che, Y, Chi, X, Qiu, X, Han, S, Dormitzer, P.R, Yang, X. | | Deposit date: | 2020-10-09 | | Release date: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Prefusion structure of human cytomegalovirus glycoprotein B and structural basis for membrane fusion.
Sci Adv, 7, 2021
|
|
7KDD
 
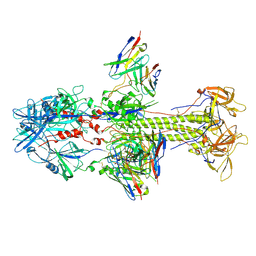 | | HCMV postfusion gB in complex with SM5-1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Envelope glycoprotein B, ... | | Authors: | Liu, Y, Heim, P.K, Che, Y, Chi, X, Qiu, X, Han, S, Dormitzer, P.R, Yang, X. | | Deposit date: | 2020-10-08 | | Release date: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Prefusion structure of human cytomegalovirus glycoprotein B and structural basis for membrane fusion.
Sci Adv, 7, 2021
|
|
5IE1
 
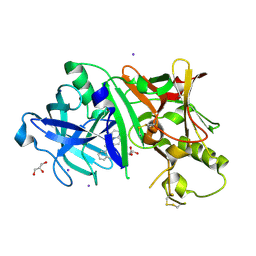 | | Crystal structure of BACE1 in complex with 3-(2-amino-6-(o-tolyl)quinolin-3-yl)-N-(3,3-dimethylbutyl)propanamide | | Descriptor: | 3-[2-amino-6-(2-methylphenyl)quinolin-3-yl]-N-(3,3-dimethylbutyl)propanamide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Jordan, J.B, Whittington, D.A, Bartberger, M.D, Sickmier, E.A, Chen, K, Cheng, Y, Judd, T. | | Deposit date: | 2016-02-24 | | Release date: | 2016-03-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Fragment-Linking Approach Using (19)F NMR Spectroscopy To Obtain Highly Potent and Selective Inhibitors of beta-Secretase.
J.Med.Chem., 59, 2016
|
|
5J3R
 
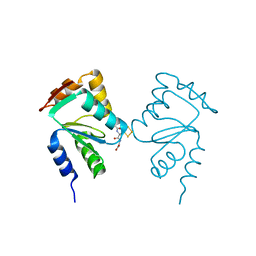 | | Crystal structure of yeast monothiol glutaredoxin Grx6 in complex with a glutathione-coordinated [2Fe-2S] cluster | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLUTATHIONE, Monothiol glutaredoxin-6 | | Authors: | Abdalla, M, Dai, Y.-N, Chi, C.-B, Cheng, W, Cao, D.-D, Zhou, K, Ali, W, Chen, Y, Zhou, C.-Z. | | Deposit date: | 2016-03-31 | | Release date: | 2016-10-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Crystal structure of yeast monothiol glutaredoxin Grx6 in complex with a glutathione-coordinated [2Fe-2S] cluster
Acta Crystallogr.,Sect.F, 72, 2016
|
|
8IOD
 
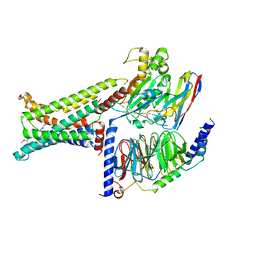 | | Cryo-EM structure of the PG-901-bound human melanocortin receptor 5 (MC5R)-Gs complex | | Descriptor: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1,HiBiT, ... | | Authors: | Feng, W.B, Zhou, Q.T, Chen, X.Y, Dai, A.T, Cai, X.Q, Liu, X, Zhao, F.H, Chen, Y, Ye, C.Y, Xu, Y.N, Cong, Z.T, Li, H, Lin, S, Yang, D.H, Wang, M.W. | | Deposit date: | 2023-03-10 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structural insights into ligand recognition and subtype selectivity of the human melanocortin-3 and melanocortin-5 receptors.
Cell Discov, 9, 2023
|
|
8INR
 
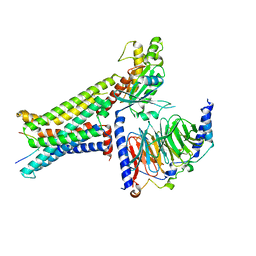 | | Cryo-EM structure of the alpha-MSH-bound human melanocortin receptor 5 (MC5R)-Gs complex | | Descriptor: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1,HiBiT, ... | | Authors: | Feng, W.B, Zhou, Q.T, Chen, X.Y, Dai, A.T, Cai, X.Q, Liu, X, Zhao, F.H, Chen, Y, Ye, C.Y, Xu, Y.N, Cong, Z.T, Li, H, Lin, S, Yang, D.H, Wang, M.W. | | Deposit date: | 2023-03-10 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural insights into ligand recognition and subtype selectivity of the human melanocortin-3 and melanocortin-5 receptors.
Cell Discov, 9, 2023
|
|
8IOC
 
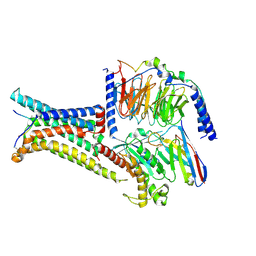 | | Cryo-EM structure of the gamma-MSH-bound human melanocortin receptor 3 (MC3R)-Gs complex | | Descriptor: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1,HiBiT, ... | | Authors: | Feng, W.B, Zhou, Q.T, Chen, X.Y, Dai, A.T, Cai, X.Q, Liu, X, Zhao, F.H, Chen, Y, Ye, C.Y, Xu, Y.N, Cong, Z.T, Li, H, Lin, S. | | Deposit date: | 2023-03-10 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural insights into ligand recognition and subtype selectivity of the human melanocortin-3 and melanocortin-5 receptors.
Cell Discov, 9, 2023
|
|
7TOB
 
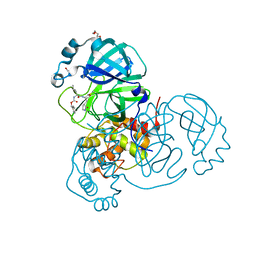 | | Crystal structure of the SARS-CoV-2 Omicron main protease (Mpro) in complex with inhibitor GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | Authors: | Sacco, M.D, Wang, J, Chen, Y. | | Deposit date: | 2022-01-24 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The P132H mutation in the main protease of Omicron SARS-CoV-2 decreases thermal stability without compromising catalysis or small-molecule drug inhibition.
Cell Res., 32, 2022
|
|
8IJR
 
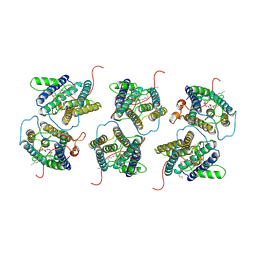 | | The cryo-EM structure of human sphingomyelin synthase-related protein in complex with diacylglycerol/phosphoethanolamine | | Descriptor: | (2S)-1-(hexadecanoyloxy)-3-hydroxypropan-2-yl (11Z)-octadec-11-enoate, PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, Sphingomyelin synthase-related protein 1 | | Authors: | Hu, K, Zhang, Q, Chen, Y, Yao, D, Zhou, L, Cao, Y. | | Deposit date: | 2023-02-28 | | Release date: | 2024-02-28 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cryo-EM structure of human sphingomyelin synthase and its mechanistic implications for sphingomyelin synthesis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8IJQ
 
 | | The cryo-EM structure of human sphingomyelin synthase-related protein in complex with ceramide | | Descriptor: | N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)PALMITAMIDE, Sphingomyelin synthase-related protein 1 | | Authors: | Hu, K, Zhang, Q, Chen, Y, Yao, D, Zhou, L, Cao, Y. | | Deposit date: | 2023-02-27 | | Release date: | 2024-02-28 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Cryo-EM structure of human sphingomyelin synthase and its mechanistic implications for sphingomyelin synthesis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
1SDI
 
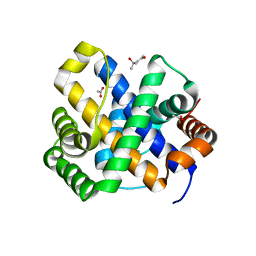 | | 1.65 A structure of Escherichia coli ycfC gene product | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, Hypothetical protein ycfC | | Authors: | Borek, D, Otwinowski, Z, Chen, Y, Skarina, T, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-13 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural analysis of Escherichia coli ycfC gene product
To be Published
|
|
7YBF
 
 | |
4Z8I
 
 | | Crystal structure of Branchiostoma belcheri tsingtauense peptidoglycan recognition protein 3 | | Descriptor: | ZINC ION, peptidoglycan recognition protein 3 | | Authors: | Wang, W.J, Cheng, W, Jiang, Y.L, Luo, M, Cao, D.D, Chi, C.B, Yang, H.B, Chen, Y, Zhou, C.Z. | | Deposit date: | 2015-04-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Activity Augmentation of Amphioxus Peptidoglycan Recognition Protein BbtPGRP3 via Fusion with a Chitin Binding Domain
Plos One, 10, 2015
|
|
3QFM
 
 | | Crystal structure of Streptococcal asymmetric Ap4A hydrolase and phosphodiesterase Spr1479/SapH | | Descriptor: | FE (III) ION, MANGANESE (II) ION, Putative uncharacterized protein | | Authors: | Jiang, Y.L, Zhang, J.W, Yu, W.L, Cheng, W, Zhang, C.C, Zhou, C.Z, Chen, Y. | | Deposit date: | 2011-01-22 | | Release date: | 2011-08-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and enzymatic characterization of a Streptococcal ATP/diadenosine polyphosphate and phosphodiester hydrolase Spr1479/SapH
To be Published
|
|
8KGR
 
 | | Structure of African swine fever virus topoisomerase II in complex with dsDNA | | Descriptor: | DNA (32-MER), DNA (33-MER), DNA topoisomerase 2, ... | | Authors: | Cong, J, Xin, U, Li, X, Chen, Y. | | Deposit date: | 2023-08-19 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of African swine fever virus topoisomerase II in complex with dsDNA
To Be Published
|
|
6EFJ
 
 | | Crystal structure of NDM-1 with compound 9 | | Descriptor: | (2R)-2-phenyl-2-(phenylamino)-N-(1H-tetrazol-5-yl)acetamide, Metallo-beta-lactamase type 2, ZINC ION | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2018-08-16 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Active-Site Druggability of Carbapenemases and Broad-Spectrum Inhibitor Discovery.
Acs Infect Dis., 5, 2019
|
|
7MZ5
 
 | |
7MZB
 
 | | Cryo-EM structure of minimal TRPV1 with 3 bound RTX and 1 perturbed PI | | Descriptor: | (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-22 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7MZ6
 
 | | Cryo-EM structure of minimal TRPV1 with 1 perturbed PI | | Descriptor: | (10R,13S)-16-amino-13-hydroxy-7,13-dioxo-8,12,14-trioxa-13lambda~5~-phosphahexadecan-10-yl hexadecanoate, (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-22 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7MZC
 
 | |
7MZA
 
 | | Cryo-EM structure of minimal TRPV1 with 2 bound RTX in adjacent pockets | | Descriptor: | (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-22 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
