7R7L
 
 | | Structure of human SHP2 in complex with compound 30 | | Descriptor: | 6-[(3S,4S)-4-amino-3-methyl-2-oxa-8-azaspiro[4.5]decan-8-yl]-3-(2,3-dichlorophenyl)-2-methylpyrimidin-4(3H)-one, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Leonard, P.G, Cross, J. | | Deposit date: | 2021-06-24 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of 6-[(3 S ,4 S )-4-Amino-3-methyl-2-oxa-8-azaspiro[4.5]decan-8-yl]-3-(2,3-dichlorophenyl)-2-methyl-3,4-dihydropyrimidin-4-one (IACS-15414), a Potent and Orally Bioavailable SHP2 Inhibitor.
J.Med.Chem., 64, 2021
|
|
7R7I
 
 | | Structure of human SHP2 in complex with compound 27 | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11, [3-(4-amino-4-methylpiperidin-1-yl)-6-(2,3-dichlorophenyl)-5-methylpyrazin-2-yl]methanol | | Authors: | Leonard, P.G, Cross, J. | | Deposit date: | 2021-06-24 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery of 6-[(3 S ,4 S )-4-Amino-3-methyl-2-oxa-8-azaspiro[4.5]decan-8-yl]-3-(2,3-dichlorophenyl)-2-methyl-3,4-dihydropyrimidin-4-one (IACS-15414), a Potent and Orally Bioavailable SHP2 Inhibitor.
J.Med.Chem., 64, 2021
|
|
5I40
 
 | | BRD9 in complex with Cpd1 (6-methyl-1,6-dihydro-7H-pyrrolo[2,3-c]pyridin-7-one) | | Descriptor: | 1,2-ETHANEDIOL, 6-methyl-1,6-dihydro-7H-pyrrolo[2,3-c]pyridin-7-one, Bromodomain-containing protein 9, ... | | Authors: | Murray, J.M. | | Deposit date: | 2016-02-11 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.0402 Å) | | Cite: | Diving into the Water: Inducible Binding Conformations for BRD4, TAF1(2), BRD9, and CECR2 Bromodomains.
J.Med.Chem., 59, 2016
|
|
4XZ7
 
 | | Crystal structure of a TGase | | Descriptor: | Putative uncharacterized protein | | Authors: | Yu, J, Ge, J, Yang, M. | | Deposit date: | 2015-02-04 | | Release date: | 2015-06-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Functional and Structural Characterization of the Antiphagocytic Properties of a Novel Transglutaminase from Streptococcus suis
J.Biol.Chem., 290, 2015
|
|
4EUF
 
 | | Crystal structure of Clostridium acetobutulicum trans-2-enoyl-CoA reductase in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative reductase CA_C0462, SODIUM ION | | Authors: | Hu, K, Zhao, M, Zhang, T, Yang, S, Ding, J. | | Deposit date: | 2012-04-25 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of trans-2-enoyl-CoA reductases from Clostridium acetobutylicum and Treponema denticola: insights into the substrate specificity and the catalytic mechanism
Biochem.J., 449, 2013
|
|
6L3F
 
 | |
6L8K
 
 | | Structure of URT1 in complex with UTP | | Descriptor: | URIDINE 5'-TRIPHOSPHATE, UTP:RNA uridylyltransferase 1 | | Authors: | Lingru, Z. | | Deposit date: | 2019-11-06 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.999 Å) | | Cite: | Crystal structure of Arabidopsis terminal uridylyl transferase URT1.
Biochem.Biophys.Res.Commun., 524, 2020
|
|
7M7D
 
 | | Crystal structure of the indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with IACS-8968 | | Descriptor: | (5S)-6,6-dimethyl-8-[(4S)-7-(trifluoromethyl)imidazo[1,5-a]pyridin-5-yl]-1,3,8-triazaspiro[4.5]decane-2,4-dione, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Leonard, P.G, Cross, J.B. | | Deposit date: | 2021-03-27 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of IACS-9779 and IACS-70465 as Potent Inhibitors Targeting Indoleamine 2,3-Dioxygenase 1 (IDO1) Apoenzyme.
J.Med.Chem., 64, 2021
|
|
7M63
 
 | | Crystal structure of the indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with IACS-70099 | | Descriptor: | (2R)-N-(4-chlorophenyl)-2-[(1R,3S,5S,6r)-3-(5,6-difluoro-1H-benzimidazol-1-yl)bicyclo[3.1.0]hexan-6-yl]propanamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Leonard, P.G, Cross, J.B. | | Deposit date: | 2021-03-25 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of IACS-9779 and IACS-70465 as Potent Inhibitors Targeting Indoleamine 2,3-Dioxygenase 1 (IDO1) Apoenzyme.
J.Med.Chem., 64, 2021
|
|
6XH7
 
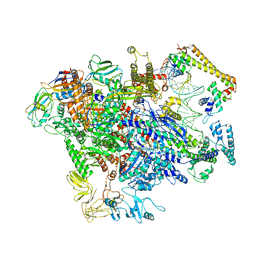 | | CueR-TAC without RNA | | Descriptor: | COPPER (II) ION, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Liu, B, Shi, W, Yang, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of copper-efflux-regulator-dependent transcription activation.
Iscience, 24, 2021
|
|
6XH8
 
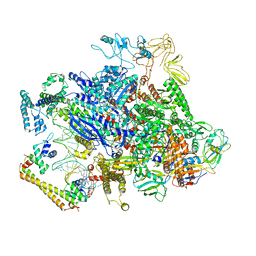 | | CueR-transcription activation complex with RNA transcript | | Descriptor: | COPPER (II) ION, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Liu, B, Shi, W, Yang, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of copper-efflux-regulator-dependent transcription activation.
Iscience, 24, 2021
|
|
6FBV
 
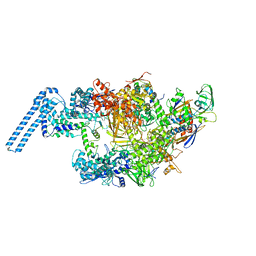 | | Single particle cryo em structure of Mycobacterium tuberculosis RNA polymerase in complex with Fidaxomicin | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Das, K, Lin, W, Ebright, E. | | Deposit date: | 2017-12-19 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structural Basis of Transcription Inhibition by Fidaxomicin (Lipiarmycin A3).
Mol. Cell, 70, 2018
|
|
8FY5
 
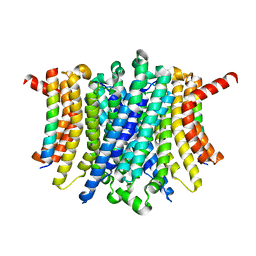 | | Human TMEM175-LAMP1 full-length complex | | Descriptor: | Endosomal/lysosomal potassium channel TMEM175, Lysosome-associated membrane glycoprotein 1 | | Authors: | Zhang, J.Y, Zeng, W.Z, Han, Y, Jiang, Y.X. | | Deposit date: | 2023-01-25 | | Release date: | 2023-06-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Lysosomal LAMP proteins regulate lysosomal pH by direct inhibition of the TMEM175 channel.
Mol.Cell, 83, 2023
|
|
8FYF
 
 | | Human TMEM175-LAMP1 transmembrane domain only complex | | Descriptor: | Endosomal/lysosomal potassium channel TMEM175, Lysosome-associated membrane glycoprotein 1 | | Authors: | Zhang, J.Y, Zeng, W.Z, Han, Y, Jiang, Y.X. | | Deposit date: | 2023-01-26 | | Release date: | 2023-06-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Lysosomal LAMP proteins regulate lysosomal pH by direct inhibition of the TMEM175 channel.
Mol.Cell, 83, 2023
|
|
5KTU
 
 | | Crystal structure of the bromodomain of human CREBBP bound to pyrazolopiperidine scaffold | | Descriptor: | 1-(3-phenylazanyl-1,4,6,7-tetrahydropyrazolo[4,3-c]pyridin-5-yl)ethanone, CREB-binding protein, DIMETHYL SULFOXIDE | | Authors: | Jayaram, H, Poy, F, Setser, J.W, Bellon, S.F. | | Deposit date: | 2016-07-12 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Discovery of a Potent and Selective in Vivo Probe (GNE-272) for the Bromodomains of CBP/EP300.
J. Med. Chem., 59, 2016
|
|
5JDM
 
 | | Structural mechanisms of extracellular ion exchange and induced binding-site occlusion in the sodium-calcium exchanger NCX_Mj soaked with 2.5 mM Na+ and 0.1mM Sr2+ | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,6,9,12,15-PENTAOXAHEPTADECANE, PENTADECANE, ... | | Authors: | Liao, J, Jiang, Y.X, Faraldo-Gomez, J.D. | | Deposit date: | 2016-04-17 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.558 Å) | | Cite: | Mechanism of extracellular ion exchange and binding-site occlusion in a sodium/calcium exchanger.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5JDH
 
 | | Structural mechanisms of extracellular ion exchange and induced binding-site occlusion in the sodium-calcium exchanger NCX_Mj soaked with 10 mM Na+ and 10mM Ca2+ | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, CALCIUM ION, ... | | Authors: | Liao, J, Jiang, Y.X, Faraldo-Gomez, J.D. | | Deposit date: | 2016-04-16 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Mechanism of extracellular ion exchange and binding-site occlusion in a sodium/calcium exchanger.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5JDN
 
 | | Structural mechanisms of extracellular ion exchange and induced binding-site occlusion in the sodium-calcium exchanger NCX_Mj soaked with 10 mM Na+ and 10mM Sr2+ | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, PENTADECANE, PENTAETHYLENE GLYCOL, ... | | Authors: | Liao, J, Jiang, Y.X, Faraldo-Gomez, J.D. | | Deposit date: | 2016-04-17 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of extracellular ion exchange and binding-site occlusion in a sodium/calcium exchanger.
Nat.Struct.Mol.Biol., 23, 2016
|
|
8CZJ
 
 | | A bacteria Zrt/Irt-like protein in the apo state | | Descriptor: | Putative membrane protein, SULFATE ION, [(Z)-octadec-9-enyl] (2R)-2,3-bis(oxidanyl)propanoate | | Authors: | Zhang, Y, Hu, J. | | Deposit date: | 2022-05-24 | | Release date: | 2023-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | Structural insights into the elevator-type transport mechanism of a bacterial ZIP metal transporter.
Nat Commun, 14, 2023
|
|
5JDG
 
 | | Structural mechanisms of extracellular ion exchange and induced binding-site occlusion in the sodium-calcium exchanger NCX_Mj soaked with 2.5 mM Na+ and 0.1mM Ca2+ | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Liao, J, Jiang, Y.X, Faraldo-Gomez, J.D. | | Deposit date: | 2016-04-16 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.407 Å) | | Cite: | Mechanism of extracellular ion exchange and binding-site occlusion in a sodium/calcium exchanger.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5JDL
 
 | | Structural mechanisms of extracellular ion exchange and induced binding-site occlusion in the sodium-calcium exchanger NCX_Mj soaked with 2.5 mM Na+ and 1mM Sr2+ | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, PENTADECANE, ... | | Authors: | Liao, J, Jiang, Y.X, Faraldo-Gomez, J.D. | | Deposit date: | 2016-04-17 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.904 Å) | | Cite: | Mechanism of extracellular ion exchange and binding-site occlusion in a sodium/calcium exchanger.
Nat.Struct.Mol.Biol., 23, 2016
|
|
4ZZJ
 
 | | SIRT1/Activator/Substrate Complex | | Descriptor: | (3S)-1,3-dimethyl-N-[3-(1,3-oxazol-5-yl)phenyl]-6-[3-(trifluoromethyl)phenyl]-2,3-dihydropyrido[2,3-b]pyrazine-4(1H)-carboxamide, Ac-p53, CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Dai, H. | | Deposit date: | 2015-05-22 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7403 Å) | | Cite: | Crystallographic structure of a small molecule SIRT1 activator-enzyme complex.
Nat Commun, 6, 2015
|
|
5JDF
 
 | | Structural mechanisms of extracellular ion exchange and induced binding-site occlusion in the sodium-calcium exchanger NCX_Mj soaked with 2.5 mM Na+ and 1mM Ca2+ | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CALCIUM ION, PENTADECANE, ... | | Authors: | Liao, J, Jiang, Y.X, Faraldo-Gomez, J.D. | | Deposit date: | 2016-04-16 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Mechanism of extracellular ion exchange and binding-site occlusion in a sodium/calcium exchanger.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5JDQ
 
 | | Structural mechanisms of extracellular ion exchange and induced binding-site occlusion in the sodium-calcium exchanger NCX_Mj soaked with 100 mM Na+ and 10mM Sr2+ | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, PENTADECANE, ... | | Authors: | Liao, J, Jiang, Y.X, Faraldo-Gomez, J.D. | | Deposit date: | 2016-04-17 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of extracellular ion exchange and binding-site occlusion in a sodium/calcium exchanger.
Nat.Struct.Mol.Biol., 23, 2016
|
|
6WU8
 
 | | Structure of human SHP2 in complex with inhibitor IACS-13909 | | Descriptor: | 1-[3-(2,3-dichlorophenyl)-1H-pyrazolo[3,4-b]pyrazin-6-yl]-4-methylpiperidin-4-amine, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Leonard, P.G, Joseph, S, Rodenberger, A. | | Deposit date: | 2020-05-04 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Allosteric SHP2 Inhibitor, IACS-13909, Overcomes EGFR-Dependent and EGFR-Independent Resistance Mechanisms toward Osimertinib.
Cancer Res., 80, 2020
|
|
