7Y5Q
 
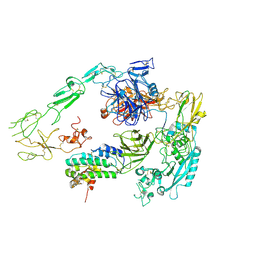 | | Structure of 1:1 PAPP-A.STC2 complex(half map) | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Pappalysin-1, Stanniocalcin-2, ZINC ION | | Authors: | Zhong, Q.H, Chu, H.L, Wang, G.P, Zhang, C, Wei, Y, Qiao, J, Hang, J. | | Deposit date: | 2022-06-17 | | Release date: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into the covalent regulation of PAPP-A activity by proMBP and STC2.
Cell Discov, 8, 2022
|
|
5YZ3
 
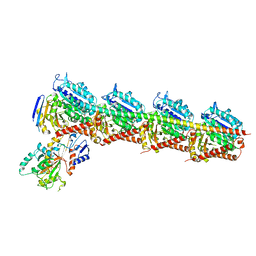 | | Crystal structure of T2R-TTL-28 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2017-12-12 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | A Novel Microtubule Inhibitor Overcomes Multidrug Resistance in Tumors.
Cancer Res., 78, 2018
|
|
8HGH
 
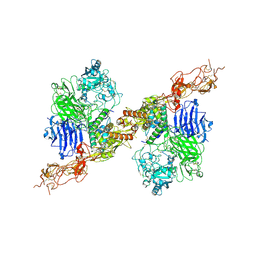 | | Structure of 2:2 PAPP-A.STC2 complex | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Pappalysin-1, Stanniocalcin-2, ZINC ION | | Authors: | Zhong, Q.H, Chu, H.L, Wang, G.P, Zhang, C, Wei, Y, Qiao, J, Hang, J. | | Deposit date: | 2022-11-14 | | Release date: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | Structural insights into the covalent regulation of PAPP-A activity by proMBP and STC2.
Cell Discov, 8, 2022
|
|
8HGG
 
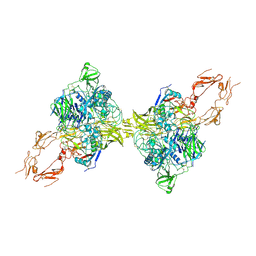 | | Structure of 2:2 PAPP-A.ProMBP complex | | Descriptor: | Bone marrow proteoglycan, Pappalysin-1, ZINC ION | | Authors: | Zhong, Q.H, Chu, H.L, Wang, G.P, Zhang, C, Wei, Y, Qiao, J, Hang, J. | | Deposit date: | 2022-11-14 | | Release date: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural insights into the covalent regulation of PAPP-A activity by proMBP and STC2.
Cell Discov, 8, 2022
|
|
1JGD
 
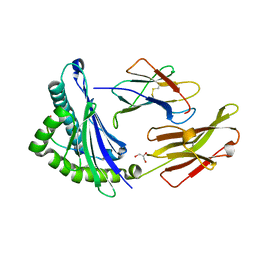 | | HLA-B*2709 bound to deca-peptide s10R | | Descriptor: | BETA-2-MICROGLOBULIN, GLYCEROL, HUMAN LYMPHOCYTE ANTIGEN HLA-B27, ... | | Authors: | Hillig, R.C, Huelsmeyer, M, Saenger, W, Volz, A, Uchanska-Ziegler, B, Ziegler, A. | | Deposit date: | 2001-06-25 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Thermodynamic and structural analysis
of peptide- and allele-dependent properties
of two HLA-B27 subtypes exhibiting differential disease
association
J.Biol.Chem., 279, 2004
|
|
7EYI
 
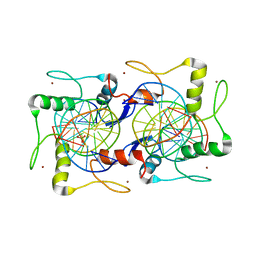 | |
6CNL
 
 | | Crystal Structure of H105A PGAM5 Dodecamer | | Descriptor: | MAGNESIUM ION, PGAM5 Multimerization Motif Peptide, Serine/threonine-protein phosphatase PGAM5, ... | | Authors: | Ruiz, K, Agnew, C, Jura, N. | | Deposit date: | 2018-03-08 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Functional role of PGAM5 multimeric assemblies and their polymerization into filaments.
Nat Commun, 10, 2019
|
|
8HNK
 
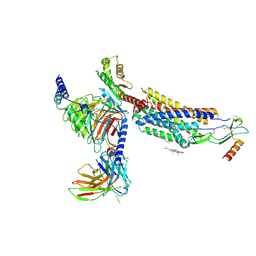 | | CXCR3-DNGi complex activated by CXCL11 | | Descriptor: | C-X-C motif chemokine 11, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Jiao, H.Z, Hu, H.L. | | Deposit date: | 2022-12-08 | | Release date: | 2023-11-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural insights into the activation and inhibition of CXC chemokine receptor 3.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HNN
 
 | | Structure of CXCR3 complexed with antagonist SCH546738 | | Descriptor: | 3-azanyl-6-chloranyl-5-[(3S)-4-[1-[(4-chlorophenyl)methyl]piperidin-4-yl]-3-ethyl-piperazin-1-yl]pyrazine-2-carboxamide, CHOLESTEROL, Nb6, ... | | Authors: | Jiao, H.Z, Hu, H.L. | | Deposit date: | 2022-12-08 | | Release date: | 2023-11-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into the activation and inhibition of CXC chemokine receptor 3.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HNL
 
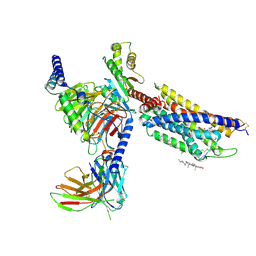 | | CXCR3-DNGi complex activated by PS372424 | | Descriptor: | (3S)-N-[(2S)-5-carbamimidamido-1-(cyclohexylmethylamino)-1-oxidanylidene-pentan-2-yl]-2-(4-oxidanylidene-4-phenyl-butanoyl)-3,4-dihydro-1H-isoquinoline-3-carboxamide, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Jiao, H.Z, Hu, H.L. | | Deposit date: | 2022-12-08 | | Release date: | 2023-11-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural insights into the activation and inhibition of CXC chemokine receptor 3.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HNM
 
 | | CXCR3-DNGi complex activated by VUF11222 | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Jiao, H.Z, Hu, H.L. | | Deposit date: | 2022-12-08 | | Release date: | 2023-11-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural insights into the activation and inhibition of CXC chemokine receptor 3.
Nat.Struct.Mol.Biol., 31, 2024
|
|
1TFX
 
 | |
1ABO
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF THE ABL TYROSINE KINASE SH3 DOMAIN WITH 3BP-1 SYNTHETIC PEPTIDE | | Descriptor: | 3BP-1 SYNTHETIC PEPTIDE, 10 RESIDUES, ABL TYROSINE KINASE, ... | | Authors: | Musacchio, A, Wilmanns, M, Saraste, M. | | Deposit date: | 1995-05-19 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution crystal structures of tyrosine kinase SH3 domains complexed with proline-rich peptides.
Nat.Struct.Biol., 1, 1994
|
|
1ABQ
 
 | |
1AWO
 
 | |
3EOR
 
 | | Crystal structure of 2C-methyl-D-erythritol 2,4-clycodiphosphate synthase complexed with ligand | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, GERANYL DIPHOSPHATE, SODIUM ION, ... | | Authors: | Hunter, W.N, Ramsden, N.L, Dawson, A. | | Deposit date: | 2008-09-29 | | Release date: | 2009-08-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A structure-based approach to ligand discovery for 2C-methyl-D-erythritol-2,4-cyclodiphosphate synthase: a target for antimicrobial therapy
J.Med.Chem., 52, 2009
|
|
3ESJ
 
 | | Crystal structure of 2C-methyl-D-erythritol 2,4-clycodiphosphate synthase complexed with ligand | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, 4-amino-1-[(2S,4aR,6R,7R,7aS)-2,7-dihydroxy-2-oxidotetrahydro-4H-furo[3,2-d][1,3,2]dioxaphosphinin-6-yl]pyrimidin-2(1H)-one, GERANYL DIPHOSPHATE, ... | | Authors: | Hunter, W.N, Ramsden, N.L. | | Deposit date: | 2008-10-06 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A structure-based approach to ligand discovery for 2C-methyl-D-erythritol-2,4-cyclodiphosphate synthase: a target for antimicrobial therapy
J.Med.Chem., 52, 2009
|
|
3ERN
 
 | | Crystal structure of 2C-methyl-D-erythritol 2,4-clycodiphosphate synthase complexed with AraCMP | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, CYTOSINE ARABINOSE-5'-PHOSPHATE, GERANYL DIPHOSPHATE, ... | | Authors: | Hunter, W.N, Ramsden, N.L, Kemp, L.A. | | Deposit date: | 2008-10-02 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structure-based approach to ligand discovery for 2C-methyl-D-erythritol-2,4-cyclodiphosphate synthase: a target for antimicrobial therapy
J.Med.Chem., 52, 2009
|
|
1SZB
 
 | | Crystal structure of the human MBL-associated protein 19 (MAp19) | | Descriptor: | CALCIUM ION, mannose binding lectin-associated serine protease-2 related protein, MAp19 (19kDa) | | Authors: | Gregory, L.A, Thielens, N.M, Arlaud, G.J, Fontecilla-Camps, J.C, Gaboriaud, C. | | Deposit date: | 2004-04-05 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The X-ray structure of human MBL-associated protein 19 (MAp19) and its interaction site with mannan-binding lectin and L-ficolin
J.Biol.Chem., 279, 2004
|
|
5W6J
 
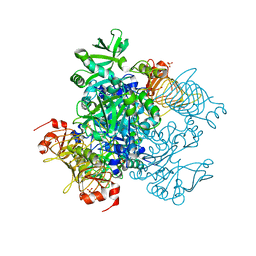 | | Agrobacterium tumefaciens ADP-glucose pyrophosphorylase | | Descriptor: | Glucose-1-phosphate adenylyltransferase, SULFATE ION | | Authors: | Mascarenhas, R.N, Hill, B.L, Wu, R, Ballicora, M.A, Liu, D. | | Deposit date: | 2017-06-16 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural analysis reveals a pyruvate-binding activator site in theAgrobacterium tumefaciensADP-glucose pyrophosphorylase.
J. Biol. Chem., 294, 2019
|
|
5W5R
 
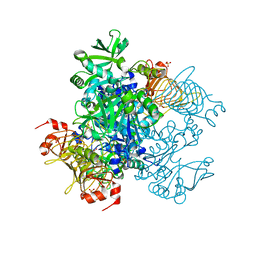 | | Agrobacterium tumefaciens ADP-glucose pyrophosphorylase P96A mutant bound to activator pyruvate | | Descriptor: | Glucose-1-phosphate adenylyltransferase, PYRUVIC ACID, SULFATE ION | | Authors: | Mascarenhas, R.N, Hill, B.L, Ballicora, M.A, Liu, D. | | Deposit date: | 2017-06-15 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.754 Å) | | Cite: | Structural analysis reveals a pyruvate-binding activator site in theAgrobacterium tumefaciensADP-glucose pyrophosphorylase.
J. Biol. Chem., 294, 2019
|
|
5W5T
 
 | | Agrobacterium tumefaciens ADP-Glucose Pyrophosphorylase bound to activator ethyl pyruvate | | Descriptor: | GLYCEROL, Glucose-1-phosphate adenylyltransferase, SULFATE ION, ... | | Authors: | Mascarenhas, R.N, Hill, B.L, Ballicora, M.A, Liu, D. | | Deposit date: | 2017-06-15 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural analysis reveals a pyruvate-binding activator site in theAgrobacterium tumefaciensADP-glucose pyrophosphorylase.
J. Biol. Chem., 294, 2019
|
|
7OLY
 
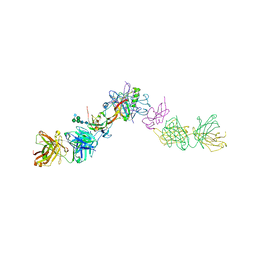 | | Structure of activin A in complex with an ActRIIB-Alk4 fusion reveal insight into activin receptor interactions | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Activin receptor type-1B, ... | | Authors: | Hakansson, M, Rose, N.C, Castonguay, R, Logan, D.T, Krishnan, L. | | Deposit date: | 2021-05-20 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.265 Å) | | Cite: | Structures of activin ligand traps using natural sets of type I and type II TGF beta receptors.
Iscience, 25, 2022
|
|
8DHK
 
 | |
8DYJ
 
 | | Crystal structure of human methylmalonyl-CoA mutase in complex with ADP and cob(II)alamin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, COBALAMIN, Methylmalonyl-CoA mutase, ... | | Authors: | Mascarenhas, R.N, Gouda, H, Banerjee, R. | | Deposit date: | 2022-08-04 | | Release date: | 2023-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bivalent molecular mimicry by ADP protects metal redox state and promotes coenzyme B 12 repair.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
