7XV9
 
 | | Crystal structure of the Human TR4 DNA-Binding Domain | | Descriptor: | Nuclear receptor subfamily 2 group C member 2, ZINC ION | | Authors: | Liu, Y, Chen, Z. | | Deposit date: | 2022-05-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Structures of human TR4LBD-JAZF1 and TR4DBD-DNA complexes reveal the molecular basis of transcriptional regulation.
Nucleic Acids Res., 51, 2023
|
|
7XV8
 
 | | Crystal structure of the Human TR4 DNA-Binding Domain Homodimer Bound to DR1 Response Element | | Descriptor: | DNA (5'-D(*CP*TP*GP*AP*CP*CP*TP*TP*TP*GP*AP*CP*CP*TP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*GP*CP*AP*GP*AP*GP*GP*TP*CP*AP*AP*AP*GP*GP*TP*CP*A)-3'), Nuclear receptor subfamily 2 group C member 2, ... | | Authors: | Liu, Y, Chen, Z. | | Deposit date: | 2022-05-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.199 Å) | | Cite: | Structures of human TR4LBD-JAZF1 and TR4DBD-DNA complexes reveal the molecular basis of transcriptional regulation.
Nucleic Acids Res., 51, 2023
|
|
7XV6
 
 | |
7XVA
 
 | |
3HK3
 
 | | Crystal structure of murine thrombin mutant W215A/E217A (one molecule in the asymmetric unit) | | Descriptor: | Thrombin heavy chain, Thrombin light chain | | Authors: | Gandhi, P.S, Page, M.J, Chen, Z, Bush-Pelc, L, Di Cera, E. | | Deposit date: | 2009-05-22 | | Release date: | 2009-07-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanism of the Anticoagulant Activity of Thrombin Mutant W215A/E217A.
J.Biol.Chem., 284, 2009
|
|
3HK6
 
 | | Crystal structure of murine thrombin mutant W215A/E217A (two molecules in the asymmetric unit) | | Descriptor: | Thrombin heavy chain, Thrombin light chain | | Authors: | Gandhi, P.S, Page, M.J, Chen, Z, Bush-Pelc, L, Di Cera, E. | | Deposit date: | 2009-05-22 | | Release date: | 2009-07-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mechanism of the Anticoagulant Activity of Thrombin Mutant W215A/E217A.
J.Biol.Chem., 284, 2009
|
|
3HKI
 
 | | Crystal structure of murine thrombin mutant W215A/E217A in complex with the extracellular fragment of human PAR1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Proteinase-activated receptor 1, Thrombin heavy chain, ... | | Authors: | Gandhi, P.S, Page, M.J, Chen, Z, Bush-Pelc, L, Di Cera, E. | | Deposit date: | 2009-05-23 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of the Anticoagulant Activity of Thrombin Mutant W215A/E217A.
J.Biol.Chem., 284, 2009
|
|
3HKJ
 
 | | Crystal structure of human thrombin mutant W215A/E217A in complex with the extracellular fragment of human PAR1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Proteinase-activated receptor 1, Thrombin heavy chain, ... | | Authors: | Gandhi, P.S, Page, M.J, Chen, Z, Bush-Pelc, L, Di Cera, E. | | Deposit date: | 2009-05-23 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanism of the Anticoagulant Activity of Thrombin Mutant W215A/E217A.
J.Biol.Chem., 284, 2009
|
|
3EDX
 
 | | Crystal structure of the W215A/E217A mutant of murine thrombin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Thrombin heavy chain, ... | | Authors: | Gandhi, P.S, Page, M.J, Chen, Z, Bush-pelc, L, Di Cera, E. | | Deposit date: | 2008-09-03 | | Release date: | 2009-07-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Basis for the Kinetic Differences of the W215A/E217A Mutant of Human and Murine Thrombin
To be Published
|
|
3EE0
 
 | | Crystal Structure of the W215A/E217A Mutant of Human Thrombin (space group P2(1)2(1)2(1)) | | Descriptor: | Thrombin heavy chain, Thrombin light chain | | Authors: | Gandhi, P.S, Page, M.J, Chen, Z, Bush-Pelc, L, Di Cera, E. | | Deposit date: | 2008-09-03 | | Release date: | 2009-07-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Molecular Basis for the Kinetic Differences of the W215A/E217A Mutant of Human and Murine Thrombin
To be Published
|
|
5EDK
 
 | | Crystal structure of prothrombin deletion mutant residues 146-167 ( Form II ). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, ... | | Authors: | Pozzi, N, Chen, Z, Di Cera, E. | | Deposit date: | 2015-10-21 | | Release date: | 2016-01-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.214 Å) | | Cite: | How the Linker Connecting the Two Kringles Influences Activation and Conformational Plasticity of Prothrombin.
J.Biol.Chem., 291, 2016
|
|
7CI1
 
 | | Crystal structure of AcrVA2 | | Descriptor: | 1,2-ETHANEDIOL, AcrVA2, SPERMIDINE | | Authors: | Chen, P, Cheng, Z, Wang, Y. | | Deposit date: | 2020-07-07 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Study on Anti-CRISPR Protein AcrVA2
Prog.Biochem.Biophys., 2021
|
|
7CI2
 
 | |
4GQH
 
 | | The Conformations and Interactions of the Four-Layer Aggregate Revealed by X-ray Crystallography Diffraction Implied the Importance of Peptides at Opposite Ends in Their Assemblies | | Descriptor: | Capsid protein | | Authors: | Li, X.Y, Song, B.A, Hu, D.Y, Chen, X, Wang, Z.C, Zeng, M.J, Yu, D.D, Chen, Z, Jin, L.H, Yang, S. | | Deposit date: | 2012-08-23 | | Release date: | 2013-08-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | The Conformations and Interactions of the Four-Layer Aggregate Revealed by X-ray Crystallography Diffraction Implied the Importance of Peptides at Opposite Ends in Their Assemblies
To be Published
|
|
4DS8
 
 | | Complex structure of abscisic acid receptor PYL3-(+)-ABA-HAB1 in the presence of Mn2+ | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3, GLYCEROL, ... | | Authors: | Zhang, X, Zhang, Q, Wang, G, Chen, Z. | | Deposit date: | 2012-02-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
8W2F
 
 | | Plasmodium falciparum 20S proteasome bound to an inhibitor | | Descriptor: | (3S)-1-[(2-fluoroethoxy)acetyl]-N-{[(4P)-4-(6-methylpyridin-3-yl)-1,3-thiazol-2-yl]methyl}piperidine-3-carboxamide, Proteasome endopeptidase complex, Proteasome subunit alpha type, ... | | Authors: | Han, Y, Deng, X, Ray, S, Chen, Z, Phillips, M. | | Deposit date: | 2024-02-20 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Identification of potent and reversible piperidine carboxamides that are species-selective orally active proteasome inhibitors to treat malaria.
Cell Chem Biol, 31, 2024
|
|
2GP9
 
 | | Crystal structure of the slow form of thrombin in a self-inhibited conformation | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Prothrombin | | Authors: | Pineda, A, Chen, Z, Mathews, F.S, Di Cera, E. | | Deposit date: | 2006-04-17 | | Release date: | 2006-09-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of thrombin in a self-inhibited conformation.
J.Biol.Chem., 281, 2006
|
|
7Y9I
 
 | | Complex structure of AtYchF1 with ppGpp | | Descriptor: | GUANOSINE-5',3'-TETRAPHOSPHATE, MAGNESIUM ION, Obg-like ATPase 1 | | Authors: | Li, X, Chen, Z. | | Deposit date: | 2022-06-24 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Co-crystalization reveals the interaction between AtYchF1 and ppGpp.
Front Mol Biosci, 9, 2022
|
|
7YDW
 
 | | Crystal structure of the MPND-DNA complex | | Descriptor: | DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), MPN domain-containing protein | | Authors: | Yang, M, Chen, Z. | | Deposit date: | 2022-07-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structures of MPND Reveal the Molecular Recognition of Nucleosomes.
Int J Mol Sci, 24, 2023
|
|
7YDT
 
 | | Crystal structure of mouse MPND | | Descriptor: | MPN domain containing protein | | Authors: | Yang, M, Chen, Z. | | Deposit date: | 2022-07-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.055 Å) | | Cite: | Structures of MPND Reveal the Molecular Recognition of Nucleosomes.
Int J Mol Sci, 24, 2023
|
|
3E6P
 
 | | Crystal structure of human meizothrombin desF1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D-PHENYLALANYL-N-[(1S)-4-{[(Z)-AMINO(IMINO)METHYL]AMINO}-1-(CHLOROACETYL)BUTYL]-L-PROLINAMIDE, Prothrombin, ... | | Authors: | Papaconstantinou, M.E, Gandhi, P, Chen, Z, Bah, A, Di Cera, E. | | Deposit date: | 2008-08-15 | | Release date: | 2008-11-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Na(+) binding to meizothrombin desF1.
Cell.Mol.Life Sci., 65, 2008
|
|
3GIC
 
 | | Structure of thrombin mutant delta(146-149e) in the free form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Thrombin heavy chain, Thrombin light chain | | Authors: | Bah, A, Carrell, C.J, Chen, Z, Gandhi, P.S, Di Cera, E. | | Deposit date: | 2009-03-05 | | Release date: | 2009-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Stabilization of the E* form turns thrombin into an anticoagulant.
J.Biol.Chem., 284, 2009
|
|
3J9K
 
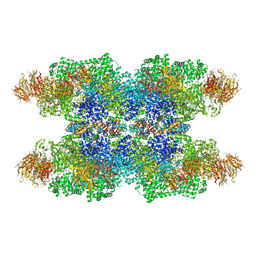 | | Structure of Dark apoptosome in complex with Dronc CARD domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Apaf-1 related killer DARK, Caspase Nc | | Authors: | Pang, Y, Bai, X, Yan, C, Hao, Q, Chen, Z, Wang, J, Scheres, S.H.W, Shi, Y. | | Deposit date: | 2015-02-04 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the apoptosome: mechanistic insights into activation of an initiator caspase from Drosophila.
Genes Dev., 29, 2015
|
|
6JDS
 
 | |
2H3X
 
 | | Crystal Structure of an Electron Transfer Complex Between Aromatic Amine Dehydrogenase and Azurin from Alcaligenes Faecalis (Form 3) | | Descriptor: | Aromatic Amine Dehydrogenase, Azurin, COPPER (II) ION | | Authors: | Sukumar, N, Chen, Z, Leys, D, Scrutton, N.S, Ferrati, D, Merli, A, Rossi, G.L, Bellamy, H.D, Chistoserdov, A, Davidson, V.L, Mathews, F.S. | | Deposit date: | 2006-05-23 | | Release date: | 2006-11-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of an Electron Transfer Complex between Aromatic Amine Dehydrogenase and Azurin from Alcaligenes faecalis.
Biochemistry, 45, 2006
|
|
