4IDV
 
 | | Crystal Structure of NIK with compound 4-{3-[2-amino-5-(2-methoxyethoxy)pyrimidin-4-yl]-1H-indol-5-yl}-2-methylbut-3-yn-2-ol (13V) | | Descriptor: | 4-{3-[2-amino-5-(2-methoxyethoxy)pyrimidin-4-yl]-1H-indol-5-yl}-2-methylbut-3-yn-2-ol, Mitogen-activated protein kinase kinase kinase 14 | | Authors: | Liu, J, Sudom, A, Wang, Z. | | Deposit date: | 2012-12-13 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Inhibiting NF-KB-inducing kinase (NIK): Discovery, structure-based design, synthesis, structure activity relationship, and co-crystal structures
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3TEF
 
 | | Crystal Structure of the Periplasmic Catecholate-Siderophore Binding Protein VctP from Vibrio Cholerae | | Descriptor: | Iron(III) ABC transporter, periplasmic iron-compound-binding protein | | Authors: | Liu, X, Wang, Z, Liu, S, Li, N, Chen, Y, Zhu, C, Zhu, D, Wei, T, Huang, Y, Xu, S, Gu, L. | | Deposit date: | 2011-08-13 | | Release date: | 2012-08-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Crystal structure of periplasmic catecholate-siderophore binding protein VctP from Vibrio cholerae at 1.7 A resolution
Febs Lett., 586, 2012
|
|
4IDT
 
 | | Crystal Structure of NIK with 11-bromo-5,6,7,8-tetrahydropyrimido[4',5':3,4]cyclohepta[1,2-b]indol-2-amine (T28) | | Descriptor: | 11-bromo-5,6,7,8-tetrahydropyrimido[4',5':3,4]cyclohepta[1,2-b]indol-2-amine, Mitogen-activated protein kinase kinase kinase 14 | | Authors: | Liu, J, Sudom, A, Wang, Z. | | Deposit date: | 2012-12-13 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibiting NF-KB-inducing kinase (NIK): Discovery, structure-based design, synthesis, structure activity relationship, and co-crystal structures
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3TGV
 
 | | Crystal structure of HutZ,the heme storsge protein from Vibrio cholerae | | Descriptor: | BENZOIC ACID, Heme-binding protein HutZ | | Authors: | Liu, X, Gong, J, Wang, Z, Du, Q, Wei, T, Zhu, D, Huang, Y, Xu, S, Gu, L. | | Deposit date: | 2011-08-17 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Crystal structure of HutZ,the heme storsge protein from Vibrio cholerae
To be Published
|
|
7LUS
 
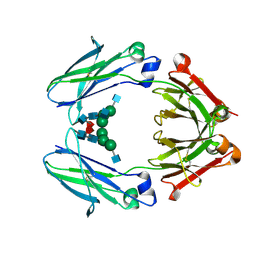 | | IgG2 Fc Charge Pair Mutation version 1 (CPMv1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin heavy constant gamma 2 | | Authors: | Sudom, A, Whittington, D, Garces, F, Wang, Z. | | Deposit date: | 2021-02-23 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Next generation Fc scaffold for multispecific antibodies.
Iscience, 24, 2021
|
|
7LUR
 
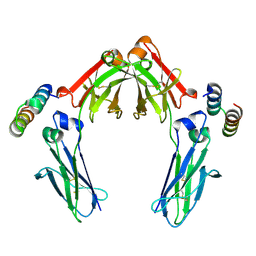 | |
4LYC
 
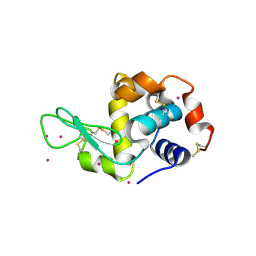 | | Cd ions within a lysoyzme single crystal | | Descriptor: | CADMIUM ION, Lysozyme C | | Authors: | Wei, H, House, S, Wu, J, Zhang, J, Wang, Z, He, Y, Gao, Y.-G, Robinson, H, Li, W, Zuo, J.-M, Robertson, I.M, Lu, Y. | | Deposit date: | 2013-07-30 | | Release date: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Enhanced and tunable fluorescent quantum dots within a single crystal of protein
TO BE PUBLISHED
|
|
8WIG
 
 | | Crystal structure of E. coli ThrS catalytic domain mutant G463S/Q484A | | Descriptor: | Threonine--tRNA ligase, ZINC ION | | Authors: | Qiao, H, Wang, Z, Wang, J, Fang, P. | | Deposit date: | 2023-09-24 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Specific glycine-dependent enzyme motion determines the potency of conformation selective inhibitors of threonyl-tRNA synthetase.
Commun Biol, 7, 2024
|
|
8WII
 
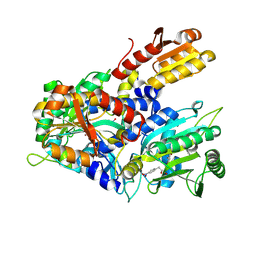 | | Crystal structure of E. coli ThrS catalytic domain mutant G463A in complex with Obafluorin | | Descriptor: | Threonine--tRNA ligase, ZINC ION, ~{N}-[(2~{R},3~{S})-2-[(4-nitrophenyl)methyl]-4-oxidanylidene-oxetan-3-yl]-2,3-bis(oxidanyl)benzamide | | Authors: | Qiao, H, Wang, Z, Wang, J, Fang, P. | | Deposit date: | 2023-09-24 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Specific glycine-dependent enzyme motion determines the potency of conformation selective inhibitors of threonyl-tRNA synthetase.
Commun Biol, 7, 2024
|
|
8WIJ
 
 | | Crystal structure of E. coli ThrS catalytic domain mutant L489M in complex with Obafluorin | | Descriptor: | N-(2,3-dihydroxybenzoyl)-4-(4-nitrophenyl)-L-threonine, Threonine--tRNA ligase, ZINC ION | | Authors: | Qiao, H, Wang, Z, Wang, J, Fang, P. | | Deposit date: | 2023-09-24 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Specific glycine-dependent enzyme motion determines the potency of conformation selective inhibitors of threonyl-tRNA synthetase.
Commun Biol, 7, 2024
|
|
5X8Y
 
 | | A Mutation identified in Neonatal Microcephaly Destabilizes Zika Virus NS1 Assembly in vitro | | Descriptor: | ZIKV NS1 | | Authors: | Wang, D, Chen, C, Liu, S, Zhou, H, Yang, K, Zhao, Q, Ji, X, Chen, C, Xie, W, Wang, Z, Mi, L.Z, Yang, H. | | Deposit date: | 2017-03-03 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.817 Å) | | Cite: | A Mutation Identified in Neonatal Microcephaly Destabilizes Zika Virus NS1 Assembly in Vitro
Sci Rep, 7, 2017
|
|
6LJG
 
 | | Crassostrea gigas ferritin mutant-D119G | | Descriptor: | FE (III) ION, Ferritin | | Authors: | Li, H, Zang, J, Tan, X, Wang, Z, Du, M. | | Deposit date: | 2019-12-15 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Crassostrea gigas ferritin mutant-D119G
To Be Published
|
|
6LIJ
 
 | | Crassostrea gigas ferritin | | Descriptor: | Ferritin, MAGNESIUM ION | | Authors: | Li, H, Zang, J, Tan, X, Wang, Z, Du, M. | | Deposit date: | 2019-12-11 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crassostrea gigas ferritin
To Be Published
|
|
6LUK
 
 | |
6LUI
 
 | | Crystal structure of the SAMD1 WH domain and DNA complex | | Descriptor: | Atherin, DNA (5'-D(*AP*CP*CP*TP*GP*CP*GP*CP*AP*CP*CP*AP*T)-3'), DNA (5'-D(*AP*TP*GP*GP*TP*GP*CP*GP*CP*AP*GP*GP*T)-3') | | Authors: | Zhou, Y, Cao, Y, Wang, Z. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-03 | | Last modified: | 2021-07-07 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | The SAM domain-containing protein 1 (SAMD1) acts as a repressive chromatin regulator at unmethylated CpG islands.
Sci Adv, 7, 2021
|
|
6LUJ
 
 | | Crystal structure of the SAMD1 SAM domain | | Descriptor: | Atherin, SULFATE ION | | Authors: | Cao, Y, Zhou, Y, Wang, Z. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | The SAM domain-containing protein 1 (SAMD1) acts as a repressive chromatin regulator at unmethylated CpG islands.
Sci Adv, 7, 2021
|
|
6MU1
 
 | | Structure of full-length IP3R1 channel bound with Adenophostin A | | Descriptor: | Adenophostin A, Inositol 1,4,5-trisphosphate receptor type 1 | | Authors: | Serysheva, I.I, Fan, G, Baker, M.R, Wang, Z, Seryshev, A, Ludtke, S.J, Baker, M.L. | | Deposit date: | 2018-10-22 | | Release date: | 2018-12-05 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM reveals ligand induced allostery underlying InsP3R channel gating.
Cell Res., 28, 2018
|
|
6BXJ
 
 | | Structure of a single-chain beta3 integrin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chimera protein of Integrin beta-3 and Integrin alpha-L, MAGNESIUM ION | | Authors: | Thinn, A.M.M, Wang, Z, Zhou, D, Zhao, Y, Curtis, B.R, Zhu, J. | | Deposit date: | 2017-12-18 | | Release date: | 2018-10-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Autonomous conformational regulation of beta3integrin and the conformation-dependent property of HPA-1a alloantibodies.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6MU2
 
 | | Structure of full-length IP3R1 channel in the Apo-state | | Descriptor: | Inositol 1,4,5-trisphosphate receptor type 1 | | Authors: | Serysheva, I.I, Fan, G, Baker, M.R, Wang, Z, Seryshev, A, Ludtke, S.J, Baker, M.L. | | Deposit date: | 2018-10-22 | | Release date: | 2018-12-05 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM reveals ligand induced allostery underlying InsP3R channel gating.
Cell Res., 28, 2018
|
|
5VT2
 
 | | Crystal structure of growth differentiation factor | | Descriptor: | 1,2-ETHANEDIOL, Growth/differentiation factor 15 | | Authors: | Min, X, Wang, Z. | | Deposit date: | 2017-05-15 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Long-acting MIC-1/GDF15 molecules to treat obesity: Evidence from mice to monkeys.
Sci Transl Med, 9, 2017
|
|
5WI9
 
 | | Crystal structure of KL with an agonist Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 39F7 Fab heavy chain, ... | | Authors: | Johnstone, S, Min, X, Wang, Z. | | Deposit date: | 2017-07-18 | | Release date: | 2018-07-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Agonistic beta-Klotho antibody mimics fibroblast growth factor 21 (FGF21) functions.
J. Biol. Chem., 293, 2018
|
|
6CNU
 
 | | Crystal Structure of JzTX-V | | Descriptor: | BROMIDE ION, GLYCEROL, JzTx-V, ... | | Authors: | Min, X, Wang, Z. | | Deposit date: | 2018-03-09 | | Release date: | 2019-03-06 | | Last modified: | 2020-03-04 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Discovery of Tarantula Venom-Derived NaV1.7-Inhibitory JzTx-V Peptide 5-Br-Trp24 Analogue AM-6120 with Systemic Block of Histamine-Induced Pruritis.
J. Med. Chem., 61, 2018
|
|
8HUD
 
 | | Cryo-EM structure of the EvCas9-sgRNA-target DNA ternary complex | | Descriptor: | CRISPR-associated endonuclease Cas9, Non-target DNA strand, Target DNA strand, ... | | Authors: | Tang, N, Wu, Z, Gao, Y, Chen, W, Su, M, Wang, Z, Ji, Q. | | Deposit date: | 2022-12-23 | | Release date: | 2023-12-27 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Molecular Basis and Genome Editing Applications of a Compact Eubacterium ventriosum CRISPR-Cas9 System.
Acs Synth Biol, 13, 2024
|
|
8GCD
 
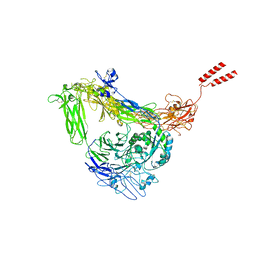 | | Full length Integrin AlphaIIbBeta3 in inactive state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Huo, T, Wu, H, Wang, Z. | | Deposit date: | 2023-03-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Full-length alpha IIb beta 3 cryo-EM structure reveals intact integrin initiate-activation intrinsic architecture.
Structure, 32, 2024
|
|
8GCE
 
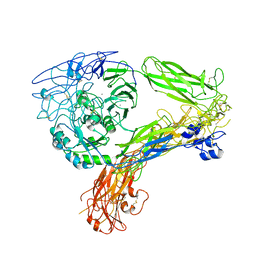 | | The Extracellular Domain of Integrin AlphaIIbBeta3 in Intermediate State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Huo, T, Wu, H, Wang, Z. | | Deposit date: | 2023-03-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Full-length alpha IIb beta 3 cryo-EM structure reveals intact integrin initiate-activation intrinsic architecture.
Structure, 32, 2024
|
|
