7PQN
 
 | | Catalytic fragment of MASP-2 in complex with ecotin | | Descriptor: | Ecotin, GLYCEROL, Mannan-binding lectin serine protease 2 A chain, ... | | Authors: | Harmat, V, Fodor, K, Heja, D. | | Deposit date: | 2021-09-17 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.400015 Å) | | Cite: | Synergy of protease-binding sites within the ecotin homodimer is crucial for inhibition of MASP enzymes and for blocking lectin pathway activation.
J.Biol.Chem., 298, 2022
|
|
8TUA
 
 | | Full-length P-Rex1 in complex with inositol 1,3,4,5-tetrakisphosphate (IP4) | | Descriptor: | INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein | | Authors: | Cash, J.N, Tesmer, J.J.G. | | Deposit date: | 2023-08-15 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Full-length P-Rex1 in complex with inositol 1,3,4,5-tetrakisphosphate (IP4)
Elife, 2024
|
|
4TNH
 
 | | RT XFEL structure of Photosystem II in the dark state at 4.9 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.900007 Å) | | Cite: | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
4TNL
 
 | | 1.8 A resolution room temperature structure of Thermolysin recorded using an XFEL | | Descriptor: | CALCIUM ION, Thermolysin, ZINC ION | | Authors: | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
8BHD
 
 | | N-terminal domain of Plasmodium berghei glutamyl-tRNA synthetase (Tbxo4 derivative crystal structure) | | Descriptor: | GLYCEROL, Glutamate--tRNA ligase, SULFATE ION, ... | | Authors: | Benas, P, Jaramillo Ponce, J.R, Legrand, P, Frugier, M, Sauter, C. | | Deposit date: | 2022-10-31 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Solution X-ray scattering highlights discrepancies in Plasmodium multi-aminoacyl-tRNA synthetase complexes.
Protein Sci., 32, 2023
|
|
6MF2
 
 | | Improved Model of Human Coagulation Factor VIII | | Descriptor: | CALCIUM ION, COPPER (I) ION, Coagulation factor VIII, ... | | Authors: | Smith, I.W, Spiegel, P.C. | | Deposit date: | 2018-09-08 | | Release date: | 2019-09-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.609364 Å) | | Cite: | The 3.2 angstrom structure of a bioengineered variant of blood coagulation factor VIII indicates two conformations of the C2 domain.
J.Thromb.Haemost., 18, 2020
|
|
7KWO
 
 | | rFVIIIFc-VWF-XTEN (BIVV001) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Fuller, J.R, Batchelor, J.D. | | Deposit date: | 2020-12-01 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular determinants of the factor VIII/von Willebrand factor complex revealed by BIVV001 cryo-electron microscopy.
Blood, 137, 2021
|
|
6U0O
 
 | | Crystal structure of a peptidoglycan release complex, SagB-SpdC, in lipidic cubic phase | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(2-ETHOXYETHOXY)ETHANOL, CITRATE ANION, ... | | Authors: | Owens, T.W, Schaefer, K, Kahne, D, Walker, S. | | Deposit date: | 2019-08-14 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and reconstitution of a hydrolase complex that may release peptidoglycan from the membrane after polymerization.
Nat Microbiol, 6, 2021
|
|
7ZAK
 
 | | Crystal structure of HLA-DP (DPA1*02:01-DPB1*01:01) in complex with a peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Racle, J, Guillaume, P, Larabi, A, Lau, K, Pojer, F, Gfeller, D. | | Deposit date: | 2022-03-22 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Machine learning predictions of MHC-II specificities reveal alternative binding mode of class II epitopes.
Immunity, 56, 2023
|
|
8FSD
 
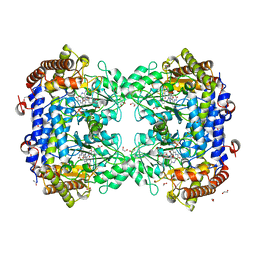 | | P130R mutant of soybean SHMT8 in complex with PLP-glycine and formylTHF | | Descriptor: | 1,2-ETHANEDIOL, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, ... | | Authors: | Beamer, L.J, Korasick, D.A. | | Deposit date: | 2023-01-09 | | Release date: | 2023-10-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural and functional analysis of two SHMT8 variants associated with soybean cyst nematode resistance.
Febs J., 291, 2024
|
|
6XA9
 
 | | SARS CoV-2 PLpro in complex with ISG15 C-terminal domain propargylamide | | Descriptor: | GLYCEROL, ISG15 CTD-propargylamide, Non-structural protein 3, ... | | Authors: | Klemm, T, Calleja, D.J, Richardson, L.W, Lechtenberg, B.C, Komander, D. | | Deposit date: | 2020-06-04 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2.
Embo J., 39, 2020
|
|
6OVT
 
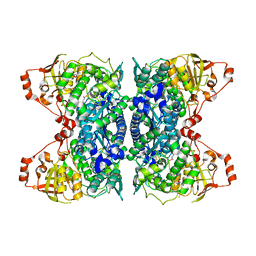 | | Crystal Structure of IlvD from Mycobacterium tuberculosis | | Descriptor: | DI(HYDROXYETHYL)ETHER, Dihydroxy-acid dehydratase, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Almo, S.C, Grove, T.L, Bonanno, J.B, Baker, E.N, Bashiri, G. | | Deposit date: | 2019-05-08 | | Release date: | 2019-08-07 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The active site of theMycobacterium tuberculosisbranched-chain amino acid biosynthesis enzyme dihydroxyacid dehydratase contains a 2Fe-2S cluster.
J.Biol.Chem., 294, 2019
|
|
7ZFR
 
 | | Crystal structure of HLA-DP (DPA1*02:01-DPB1*01:01) in complex with a peptide bound in the reverse direction | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MHC class II HLA-DP alpha chain (DPA1*02:01), MHC class II HLA-DP beta chain (DPB1*01:01), ... | | Authors: | Racle, J, Guillaume, P, Larabi, A, Lau, K, Pojer, F, Gfeller, D. | | Deposit date: | 2022-04-01 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Machine learning predictions of MHC-II specificities reveal alternative binding mode of class II epitopes.
Immunity, 56, 2023
|
|
6OPJ
 
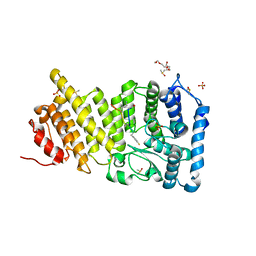 | | Menin in complex with peptide inhibitor 25 | | Descriptor: | DIMETHYL SULFOXIDE, Menin, Peptide inhibitor 25, ... | | Authors: | Linhares, B.M, Fortuna, P, Cierpicki, T, Grembecka, J, Berlicki, L. | | Deposit date: | 2019-04-25 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5006572 Å) | | Cite: | Covalent and noncovalent constraints yield a figure eight-like conformation of a peptide inhibiting the menin-MLL interaction.
Eur.J.Med.Chem., 207, 2020
|
|
8ERX
 
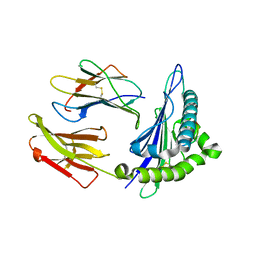 | | Structure of chimeric HLA-A*11:01-A*02:01 bound to HIV-1 RT peptide | | Descriptor: | Beta-2-microglobulin, HIV-1 RT, HLA-A*02:01 | | Authors: | Florio, T.J, Ani, O, Young, M.C, Mallik, L, Sgourakis, N.G. | | Deposit date: | 2022-10-13 | | Release date: | 2023-01-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Decoupling peptide binding from T cell receptor recognition with engineered chimeric MHC-I molecules.
Front Immunol, 14, 2023
|
|
6U1V
 
 | | Crystal structure of acyl-ACP/acyl-CoA dehydrogenase from allylmalonyl-CoA and FK506 biosynthesis, TcsD | | Descriptor: | Acyl-CoA dehydrogenase domain-containing protein, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE | | Authors: | Blake-Hedges, J.M, Pereira, J.H, Barajas, J.F, Adams, P.D, Keasling, J.D. | | Deposit date: | 2019-08-16 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Mechanism of Regioselectivity in an Unusual Bacterial Acyl-CoA Dehydrogenase.
J.Am.Chem.Soc., 142, 2020
|
|
8EGK
 
 | | Re-refinement of Crystal Structure of NosGet3d, the All4481 protein from Nostoc sp. PCC 7120 | | Descriptor: | AZIDE ION, All4481 protein | | Authors: | Barlow, A.N, Manu, M.S, Ramasamy, S, Clemons Jr, W.M. | | Deposit date: | 2022-09-12 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structures of Get3d reveal a distinct architecture associated with the emergence of photosynthesis.
J.Biol.Chem., 299, 2023
|
|
7LM4
 
 | | The crystal structure of the I38T mutant PA Endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000988503 | | Descriptor: | 5-hydroxy-N-[2-(4-hydroxy-3-methoxyphenyl)ethyl]-2-(2-methylphenyl)-6-oxo-1,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Jayaraman, S, Rankovic, Z, White, S.W. | | Deposit date: | 2021-02-05 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
6PHJ
 
 | |
6PHO
 
 | |
6P0J
 
 | | Crystal structure of GDP-bound human RalA | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Ral-A | | Authors: | Bum-Erdene, K, Gonzalez-Gutierrez, G, Liu, D, Meroueh, S.O. | | Deposit date: | 2019-05-17 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Small-molecule covalent bond formation at tyrosine creates a binding site and inhibits activation of Ral GTPases.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6PHK
 
 | |
6PHP
 
 | |
6PQ7
 
 | |
6PHN
 
 | |
