7VM4
 
 | | Crystal structure of uPA in complex with nafamostat | | Descriptor: | 4-carbamimidamidobenzoic acid, TRIETHYLENE GLYCOL, Urokinase-type plasminogen activator | | Authors: | Jiang, L.G, Huang, M.D. | | Deposit date: | 2021-10-07 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural study of the uPA-nafamostat complex reveals a covalent inhibitory mechanism of nafamostat.
Biophys.J., 121, 2022
|
|
7VM5
 
 | |
7VM7
 
 | | Crystal structure of inactive uPA in complex with nafamostat | | Descriptor: | (6-carbamimidoylnaphthalen-2-yl) 4-carbamimidamidobenzoate, Urokinase-type plasminogen activator chain B | | Authors: | Jiang, L.G, Huang, M.D. | | Deposit date: | 2021-10-07 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural study of the uPA-nafamostat complex reveals a covalent inhibitory mechanism of nafamostat.
Biophys.J., 121, 2022
|
|
6ITE
 
 | | Crystal structure of group A Streptococcal surface dehydrogenase (SDH) | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Yuan, C, Li, R, Huang, M.D. | | Deposit date: | 2018-11-21 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.739 Å) | | Cite: | Structural determination of group A Streptococcal surface dehydrogenase and characterization of its interaction with urokinase-type plasminogen activator receptor.
Biochem.Biophys.Res.Commun., 510, 2019
|
|
5W0X
 
 | | Crystal structure of mouse TOR signaling pathway regulator-like (TIPRL) delta 94-103 | | Descriptor: | TIP41-like protein | | Authors: | Wu, C, Zheng, A, Li, J, Satyshur, K, Xing, Y. | | Deposit date: | 2017-06-01 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.717 Å) | | Cite: | Methylation-regulated decommissioning of multimeric PP2A complexes.
Nat Commun, 8, 2017
|
|
8VDJ
 
 | | Crystal structure of SARS-CoV-2 3CL protease (3CLpro) as a covalent complex with EDP-235 | | Descriptor: | 3C-like proteinase nsp5, 4,6,7-trifluoro-N-{(2S)-1-[(3R,5'R)-5'-(iminomethyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidin]-1'-yl]-4-methyl-1-oxopentan-2-yl}-N-methyl-1H-indole-2-carboxamide, THIOCYANATE ION | | Authors: | Cade, I.A, Rhodin, M.H.J. | | Deposit date: | 2023-12-15 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | The small molecule inhibitor of SARS-CoV-2 3CLpro EDP-235 prevents viral replication and transmission in vivo.
Nat Commun, 15, 2024
|
|
5W0W
 
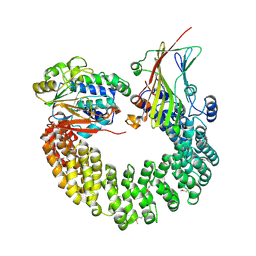 | | Crystal structure of Protein Phosphatase 2A bound to TIPRL | | Descriptor: | MANGANESE (II) ION, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, Serine/threonine-protein phosphatase 2A catalytic subunit alpha isoform, ... | | Authors: | Wu, C, Zheng, A, Li, J, Satyshur, K, Xing, Y. | | Deposit date: | 2017-06-01 | | Release date: | 2018-01-17 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Methylation-regulated decommissioning of multimeric PP2A complexes.
Nat Commun, 8, 2017
|
|
7VM6
 
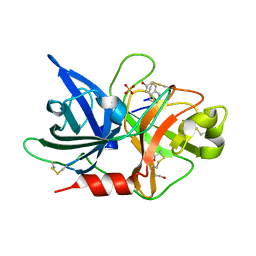 | | Crystal structure of uPA in complex with 6-amidino-2-naphthol | | Descriptor: | 6-oxidanylnaphthalene-2-carboximidamide, SULFATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Jiang, L.G, Huang, M.D. | | Deposit date: | 2021-10-07 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural study of the uPA-nafamostat complex reveals a covalent inhibitory mechanism of nafamostat.
Biophys.J., 121, 2022
|
|
8JI8
 
 | | Crystal Structure of Prophenoloxidase PPO6 chimeric mutant (F215EASNRAIVD224 to G215DGPDSVVR223) from Aedes aegypti | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COPPER (II) ION, ... | | Authors: | Zhu, X, Zhang, L, Yang, X, Bao, P, Ren, D, Han, Q. | | Deposit date: | 2023-05-26 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Mosquitoes have evolved two types of prophenoloxidases
To Be Published
|
|
8JIB
 
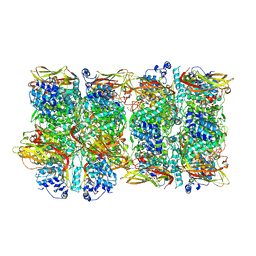 | | Crystal Structure of Prophenoloxidase PPO6 from Aedes aegypti | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, TK receptor | | Authors: | Zhu, X, Zhang, L, Yang, X, Bao, P, Ren, D, Han, Q. | | Deposit date: | 2023-05-26 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Mosquitoes have evolved two types of prophenoloxidases
To Be Published
|
|
8Y6F
 
 | |
8HLM
 
 | | Crystal structure of p53/BCL2 fusion complex (complex 2) | | Descriptor: | Apoptosis regulator Bcl-2, Cellular tumor antigen p53, ZINC ION | | Authors: | Guo, M, Wang, H, Wei, H, Chen, Y. | | Deposit date: | 2022-11-30 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.522 Å) | | Cite: | Structures of p53/BCL-2 complex suggest a mechanism for p53 to antagonize BCL-2 activity.
Nat Commun, 14, 2023
|
|
8HLL
 
 | | Crystal structure of p53/BCL2 fusion complex (complex 1) | | Descriptor: | Apoptosis regulator Bcl-2, Cellular tumor antigen p53, ZINC ION | | Authors: | Wei, H, Guo, M, Wang, H, Chen, Y. | | Deposit date: | 2022-11-30 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structures of p53/BCL-2 complex suggest a mechanism for p53 to antagonize BCL-2 activity.
Nat Commun, 14, 2023
|
|
8HLN
 
 | | Crystal structure of p53/BCL2 fusion complex(complex3) | | Descriptor: | Apoptosis regulator Bcl-2, Cellular tumor antigen p53, ZINC ION | | Authors: | Guo, M, Wei, H, Wang, H, Chen, Y. | | Deposit date: | 2022-11-30 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.354 Å) | | Cite: | Structures of p53/BCL-2 complex suggest a mechanism for p53 to antagonize BCL-2 activity.
Nat Commun, 14, 2023
|
|
5Z9R
 
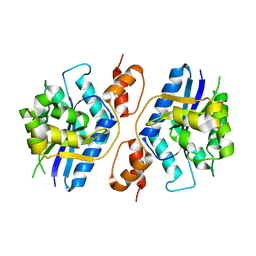 | |
7D57
 
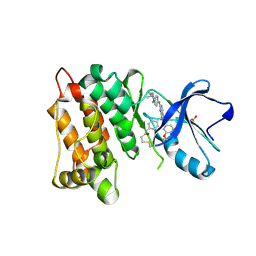 | | C-Src in complex with FIIN-2 | | Descriptor: | N-(4-{[3-(3,5-dimethoxyphenyl)-7-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-2-oxo-3,4-dihydropyrimido[4,5-d]pyrimidin-1(2H)-yl]methyl}phenyl)propanamide, Proto-oncogene tyrosine-protein kinase Src, SULFATE ION | | Authors: | Chen, Y.H, Qu, L.Z. | | Deposit date: | 2020-09-25 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Structural insights into the potency and selectivity of covalent pan-FGFR inhibitors
Commun Chem, 5, 2022
|
|
7D5O
 
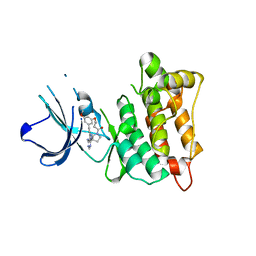 | | C-Src in complex with TAS-120 | | Descriptor: | 1-[(3S)-3-{4-amino-3-[(3,5-dimethoxyphenyl)ethynyl]-1H-pyrazolo[3,4-d]pyrimidin-1-yl}pyrrolidin-1-yl]prop-2-en-1-one, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Qu, L.Z, Chen, Y.H. | | Deposit date: | 2020-09-27 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural insights into the potency and selectivity of covalent pan-FGFR inhibitors
Commun Chem, 5, 2022
|
|
7DHT
 
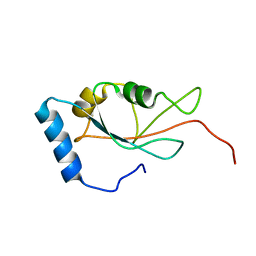 | |
7E17
 
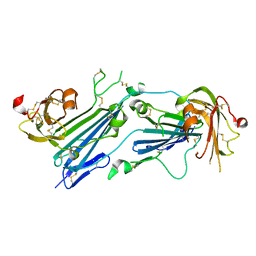 | | Structure of dimeric uPAR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Urokinase plasminogen activator surface receptor | | Authors: | Cai, Y, Huang, M. | | Deposit date: | 2021-02-01 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Crystal structure and cellular functions of uPAR dimer
Nat Commun, 13, 2022
|
|
5ZHH
 
 | |
7XUZ
 
 | | Crystal structure of a HDAC4-MEF2A-DNA ternary complex | | Descriptor: | DNA (5'-D(*TP*CP*TP*TP*AP*TP*AP*AP*AP*TP*AP*GP*T)-3'), DNA (5'-D(P*AP*CP*TP*AP*TP*TP*TP*AP*TP*AP*A)-3'), Histone deacetylase 4, ... | | Authors: | Dai, S.Y, Guo, L, Dey, R, Guo, M, Bates, D, Cayford, J, Chen, X.J, Wei, X.D, Chen, L, Chen, Y.H. | | Deposit date: | 2022-05-20 | | Release date: | 2023-11-29 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3.591 Å) | | Cite: | Structural insights into the HDAC4-MEF2A-DNA complex and its implication in long-range transcriptional regulation.
Nucleic Acids Res., 52, 2024
|
|
7F3M
 
 | | Crystal structure of FGFR4 kinase domain with PRN1371 | | Descriptor: | 6-[2,6-bis(chloranyl)-3,5-dimethoxy-phenyl]-2-(methylamino)-8-[3-(4-prop-2-enoylpiperazin-1-yl)propyl]pyrido[2,3-d]pyrimidin-7-one, Fibroblast growth factor receptor 4, SULFATE ION | | Authors: | Chen, X.J, Qu, L.Z, Dai, S.Y, Wei, H.D, Chen, Y.H. | | Deposit date: | 2021-06-16 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.289 Å) | | Cite: | Structural insights into the potency and selectivity of covalent pan-FGFR inhibitors
Commun Chem, 5, 2022
|
|
6AZR
 
 | | Crystal structure of the T264A HK853cp-BeF3-RR468 complex | | Descriptor: | Chemotaxis regulator-transmits chemoreceptor signals to flagelllar motor components CheY, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Rose, J, Zhou, P. | | Deposit date: | 2017-09-11 | | Release date: | 2017-12-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.628 Å) | | Cite: | A pH-gated conformational switch regulates the phosphatase activity of bifunctional HisKA-family histidine kinases.
Nat Commun, 8, 2017
|
|
7XV9
 
 | | Crystal structure of the Human TR4 DNA-Binding Domain | | Descriptor: | Nuclear receptor subfamily 2 group C member 2, ZINC ION | | Authors: | Liu, Y, Chen, Z. | | Deposit date: | 2022-05-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Structures of human TR4LBD-JAZF1 and TR4DBD-DNA complexes reveal the molecular basis of transcriptional regulation.
Nucleic Acids Res., 51, 2023
|
|
7XV8
 
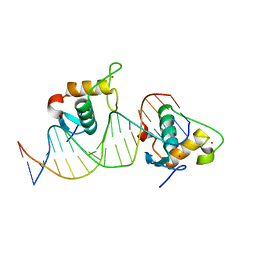 | | Crystal structure of the Human TR4 DNA-Binding Domain Homodimer Bound to DR1 Response Element | | Descriptor: | DNA (5'-D(*CP*TP*GP*AP*CP*CP*TP*TP*TP*GP*AP*CP*CP*TP*CP*TP*GP*C)-3'), DNA (5'-D(*GP*GP*CP*AP*GP*AP*GP*GP*TP*CP*AP*AP*AP*GP*GP*TP*CP*A)-3'), Nuclear receptor subfamily 2 group C member 2, ... | | Authors: | Liu, Y, Chen, Z. | | Deposit date: | 2022-05-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.199 Å) | | Cite: | Structures of human TR4LBD-JAZF1 and TR4DBD-DNA complexes reveal the molecular basis of transcriptional regulation.
Nucleic Acids Res., 51, 2023
|
|
