7VYQ
 
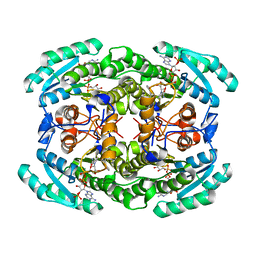 | | Short chain dehydrogenase (SCR) cryoEM structure with NADP and ethyl 4-chloroacetoacetate | | Descriptor: | Carbonyl Reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ethyl 4-chloranyl-3-oxidanylidene-butanoate | | Authors: | Li, Y.H, Zhang, R.Z, Wang, C, Forouhar, F, Clarke, O, Vorobiev, S, Singh, S, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | Deposit date: | 2021-11-15 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
3SWV
 
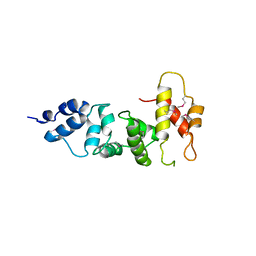 | | Crystal Structure of a domain of Brefeldin A-inhibited guanine nucleotide-exchange protein 2 (Brefeldin A-inhibited GEP 2) from Homo sapiens (Human), Northeast Structural Genomics Consortium target id HR5562A | | Descriptor: | Brefeldin A-inhibited guanine nucleotide-exchange protein 2 | | Authors: | Seetharaman, J, Su, M, Forouhar, F, Wang, D, Janjua, H, Cunningham, K, Owens, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-07-14 | | Release date: | 2011-08-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of a domain of Brefeldin A-inhibited guanine nucleotide-exchange protein 2 (BrefeldinA-inhibited GEP 2) from Homo sapiens (Human), Northeast Structural Genomics Consortium target id HR5562A
To be Published
|
|
3THT
 
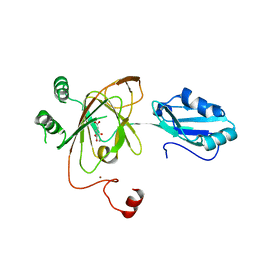 | | Crystal structure and RNA binding properties of the RRM/AlkB domains in human ABH8, an enzyme catalyzing tRNA hypermodification, Northeast Structural Genomics Consortium Target HR5601B | | Descriptor: | 2-OXOGLUTARIC ACID, Alkylated DNA repair protein alkB homolog 8, MANGANESE (II) ION, ... | | Authors: | Pastore, C, Topalidou, I, Forouhar, F, Yan, A.C, Levy, M, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-08-19 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal structure and RNA binding properties of the RNA recognition motif (RRM) and AlkB domains in human AlkB homolog 8 (ABH8), an enzyme catalyzing tRNA hypermodification.
J.Biol.Chem., 287, 2012
|
|
3THP
 
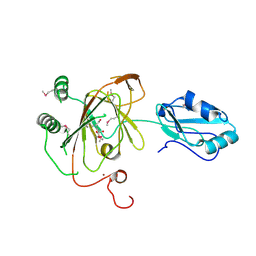 | | Crystal structure and RNA binding properties of the RRM/AlkB domains in human ABH8, an enzyme catalyzing tRNA hypermodification, Northeast Structural Genomics Consortium Target HR5601B | | Descriptor: | 2-OXOGLUTARIC ACID, Alkylated DNA repair protein alkB homolog 8, MANGANESE (II) ION, ... | | Authors: | Pastore, C, Topalidou, I, Forouhar, F, Yan, A.C, Levy, M, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-08-19 | | Release date: | 2011-11-02 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure and RNA binding properties of the RNA recognition motif (RRM) and AlkB domains in human AlkB homolog 8 (ABH8), an enzyme catalyzing tRNA hypermodification.
J.Biol.Chem., 287, 2012
|
|
7JST
 
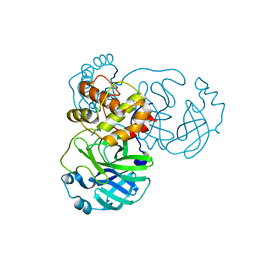 | | Crystal structure of SARS-CoV-2 3CL in apo form | | Descriptor: | 3C-like proteinase, PHOSPHATE ION | | Authors: | Iketani, S, Forouhar, F, Liu, H, Hong, S.J, Lin, F.-Y, Nair, M.S, Zask, A, Huang, Y, Xing, L, Stockwell, B.R, Chavez, A, Ho, D.D. | | Deposit date: | 2020-08-16 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Lead compounds for the development of SARS-CoV-2 3CL protease inhibitors.
Nat Commun, 12, 2021
|
|
7JT0
 
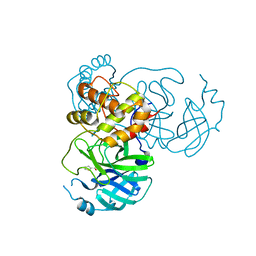 | | Crystal structure of SARS-CoV-2 3CL protease in complex with MAC5576 | | Descriptor: | 3C-like proteinase, PHOSPHATE ION, thiophene-2-carbaldehyde | | Authors: | Iketani, S, Forouhar, F, Liu, H, Hong, S.J, Lin, F.-Y, Nair, M.S, Zask, A, Huang, Y, Xing, L, Stockwell, B.R, Chavez, A, Ho, D.D. | | Deposit date: | 2020-08-16 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Lead compounds for the development of SARS-CoV-2 3CL protease inhibitors.
Nat Commun, 12, 2021
|
|
7JT7
 
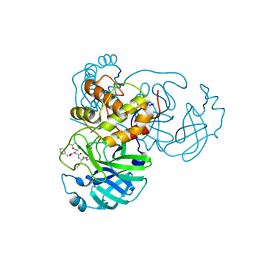 | | Crystal structure of SARS-CoV-2 3CL protease in complex with compound 4 | | Descriptor: | 3C-like proteinase, ethyl (4R)-4-[[(2S)-4-methyl-2-[[(2S,3R)-3-[(2-methylpropan-2-yl)oxy]-2-(phenylmethoxycarbonylamino)butanoyl]amino]pentanoyl]amino]-5-[(3S)-2-oxidanylidenepyrrolidin-3-yl]pentanoate | | Authors: | Iketani, S, Forouhar, F, Liu, H, Hong, S.J, Lin, F.-Y, Nair, M.S, Zask, A, Huang, Y, Xing, L, Stockwell, B.R, Chavez, A, Ho, D.D. | | Deposit date: | 2020-08-17 | | Release date: | 2021-03-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Lead compounds for the development of SARS-CoV-2 3CL protease inhibitors.
Nat Commun, 12, 2021
|
|
7JW8
 
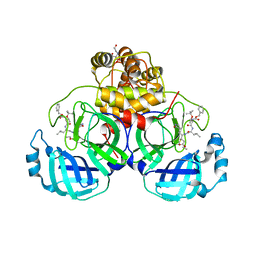 | | Crystal structure of SARS-CoV-2 3CL protease in complex with compound 4 in space group P1 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3C-like proteinase, ethyl (4R)-4-[[(2S)-4-methyl-2-[[(2S,3R)-3-[(2-methylpropan-2-yl)oxy]-2-(phenylmethoxycarbonylamino)butanoyl]amino]pentanoyl]amino]-5-[(3S)-2-oxidanylidenepyrrolidin-3-yl]pentanoate | | Authors: | Iketani, S, Forouhar, F, Liu, H, Hong, S.J, Lin, F.-Y, Nair, M.S, Zask, A, Xing, L, Stockwell, B.R, Chavez, A, Ho, D.D. | | Deposit date: | 2020-08-25 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Lead compounds for the development of SARS-CoV-2 3CL protease inhibitors.
Nat Commun, 12, 2021
|
|
7JSU
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with GC376 | | Descriptor: | 3C-like proteinase, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide, PHOSPHATE ION | | Authors: | Iketani, S, Forouhar, F, Liu, H, Hong, S.J, Lin, F.-Y, Nair, M.S, Zask, A, Xing, L, Stockwell, B.R, Chavez, A, Ho, D.D. | | Deposit date: | 2020-08-16 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Lead compounds for the development of SARS-CoV-2 3CL protease inhibitors.
Nat Commun, 12, 2021
|
|
2KW2
 
 | | Solution NMR of the specialized acyl carrier protein (RPA2022) from Rhodopseudomonas palustris, Northeast Structural Genomics Consortium Target RpR324 | | Descriptor: | Specialized acyl carrier protein | | Authors: | Rossi, P, Lee, H, Wohlbold, T.J, Valafar, H, Lemak, A, Ertekin, A, Forouhar, F, Ciccosanti, C, Rost, B, Acton, T, Xiao, R, Everett, J, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-30 | | Release date: | 2010-04-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of a specialized acyl carrier protein essential for lipid A biosynthesis with very long-chain fatty acids in open and closed conformations.
Biochemistry, 51, 2012
|
|
3Q69
 
 | | X-ray crystal structure of a MucBP domain of the protein LBA1460 from Lactobacillus acidophilus, Northeast structural genomics consortium target LaR80A | | Descriptor: | probable Zn binding protein | | Authors: | Seetharaman, J, Lew, S, Forouhar, F, Wang, D, Ciccosanti, C, Sahdev, S, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-12-31 | | Release date: | 2011-04-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystal structure of a MucBP domain of the protein LBA1460 from Lactobacillus acidophilus, Northeast structural genomics consortium target LaR80A
To be Published
|
|
2IDB
 
 | | Crystal Structure of 3-octaprenyl-4-hydroxybenzoate decarboxylase (UbiD) from Escherichia coli, Northeast Structural Genomics Target ER459. | | Descriptor: | 1,2-ETHANEDIOL, 3-octaprenyl-4-hydroxybenzoate carboxy-lyase, PENTAETHYLENE GLYCOL | | Authors: | Zhou, W, Forouhar, F, Seetharaman, J, Fang, Y, Xiao, R, Cunningham, K, Ma, L.-C, Chen, C.X, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-09-14 | | Release date: | 2006-10-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of 3-octaprenyl-4-hydroxybenzoate decarboxylase (UbiD) from Escherichia coli, Northeast Structural Genomics Target ER459.
TO BE PUBLISHED
|
|
2NMM
 
 | | Crystal structure of human phosphohistidine phosphatase. Northeast Structural Genomics Consortium target HR1409 | | Descriptor: | 14 kDa phosphohistidine phosphatase, SULFATE ION | | Authors: | Kuzin, A.P, Abashidze, M, Forouhar, F, Seetharaman, J, Kent, C, Fang, Y, Cunningham, K, Conover, K, Ma, L.C, Xiao, R, Acton, T, Montelione, G, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-10-22 | | Release date: | 2007-01-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human phosphohistidine phosphatase. Northeast Structural Genomics Consortium target HR1409
To be Published
|
|
1SQH
 
 | | X-RAY STRUCTURE OF DROSOPHILA MALONOGASTER PROTEIN Q9VR51 NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET FR87. | | Descriptor: | hypothetical protein CG14615-PA | | Authors: | Kuzin, A.P, Chen, Y, Forouhar, F, Acton, T.B, Xiao, R, Cooper, B, Ho, C.K, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-03-18 | | Release date: | 2004-04-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-RAY STRUCTURE OF DROSOPHILA MALONOGASTER PROTEIN Q9VR51 NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET FR87
To be Published
|
|
1T5Y
 
 | | Crystal Structure of Northeast Structural Genomics Consortium Target HR2118: A Human Homolog of Saccharomyces cerevisiae Nip7p | | Descriptor: | SACCHAROMYCES CEREVISIAE Nip7p HOMOLOG | | Authors: | Kuzin, A.P, Chen, Y, Forouhar, F, Acton, T.B, Shastry, R, Ma, L.-C, Cooper, B, Xiao, R, Montelione, G, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-05-05 | | Release date: | 2004-06-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Northeast Structural Genomics Consortium Target HR2118:
A Human Homolog of Saccharomyces cerevisiae Nip7p
To be Published
|
|
1T3H
 
 | | X-ray Structure of Dephospho-CoA Kinase from E. coli Norteast Structural Genomics Consortium Target ER57 | | Descriptor: | Dephospho-CoA kinase, SULFATE ION | | Authors: | Kuzin, A.P, Chen, Y, Forouhar, F, Edstrom, W, Benach, J, Vorobiev, S, Acton, T, Shastry, R, Ma, L.-C, Xia, R, Montelione, G, Tong, L, Hunt, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-04-26 | | Release date: | 2004-05-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray Structure of Dephospho-CoA Kinase from E. coli
Norteast Structural Genomics Consortium Target ER57
TO BE PUBLISHED
|
|
1SBK
 
 | | X-RAY STRUCTURE OF YDII_ECOLI NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ER29. | | Descriptor: | Hypothetical protein ydiI, SULFATE ION | | Authors: | Kuzin, A.P, Edstrom, W, Vorobiev, S.M, Lee, I, Forouhar, F, Ma, L, Chiang, Y, Rong, X, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-02-10 | | Release date: | 2004-02-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray Structure of YDII_ECOLI Northeast Structural Genomics Consortium Target ER29
To be Published
|
|
1TIY
 
 | | X-RAY STRUCTURE OF GUANINE DEAMINASE FROM BACILLUS SUBTILIS NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET SR160 | | Descriptor: | Guanine deaminase, ZINC ION | | Authors: | Kuzin, A.P, Vorobiev, S, Edstrom, W, Forouhar, F, Acton, T, Shastry, R, Ma, L.-C, Chiang, Y.-W, Montelione, G, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-06-02 | | Release date: | 2004-06-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-RAY STRUCTURE OF GUANINE DEAMINASE FROM BACILLUS SUBTILIS
NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET SR160
To be published
|
|
1WYZ
 
 | | X-Ray structure of the putative methyltransferase from Bacteroides thetaiotaomicron VPI-5482 at the resolution 2.5 A. Norteast Structural Genomics Consortium target Btr28 | | Descriptor: | putative S-adenosylmethionine-dependent methyltransferase | | Authors: | Kuzin, A.P, Chen, Y, Forouhar, F, Vorobiev, S.M, Acton, T, Ma, L.-C, Xiao, R, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-02-21 | | Release date: | 2005-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-Ray structure of the putative methyltransferase from Bacteroides thetaiotaomicron VPI-5482 at the resolution 2.5 A. Norteast Structural Genomics Consortium target Btr28
To be Published
|
|
3EQE
 
 | | Crystal structure of the YubC protein from Bacillus subtilis. Northeast Structural Genomics Consortium target SR112. | | Descriptor: | FE (III) ION, Putative cystein dioxygenase | | Authors: | Vorobiev, S.M, Su, M, Seetharaman, J, Benach, J, Forouhar, F, Clayton, G, Cooper, B, Wang, H, Foote, E.L, Ciccosanti, C, Mao, L, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-09-30 | | Release date: | 2008-10-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Crystal structure of the YubC protein from Bacillus subtilis.
To be Published
|
|
3FHW
 
 | | Crystal structure of the protein priB from Bordetella parapertussis. Northeast Structural Genomics Consortium target BpR162. | | Descriptor: | DI(HYDROXYETHYL)ETHER, Primosomal replication protein n, SODIUM ION | | Authors: | Kuzin, A.P, Neely, H, Seetharaman, J, Forouhar, F, Wang, D, Mao, L, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-10 | | Release date: | 2008-12-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the protein priB from Bordetella parapertussis. Northeast Structural Genomics Consortium target BpR162.
To be Published
|
|
3FLH
 
 | | Crystal structure of lp_1913 protein from Lactobacillus plantarum,Northeast Structural Genomics Consortium Target LpR140B | | Descriptor: | BROMIDE ION, uncharacterized protein lp_1913 | | Authors: | Seetharaman, J, Chen, Y, Forouhar, F, Sahdev, S, Foote, E.L, Ciccosanti, C, Belote, R.L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-18 | | Release date: | 2009-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of lp_1913 protein from Lactobacillus plantarum,Northeast Structural Genomics Consortium Target LpR140B
To be Published
|
|
3FDF
 
 | | Crystal structure of the serine phosphatase of RNA polymerase II CTD (SSU72 superfamily) from Drosophila melanogaster. Orthorhombic crystal form. Northeast Structural Genomics Consortium target FR253. | | Descriptor: | FR253 | | Authors: | Kuzin, A.P, Chen, Y, Seetharaman, J, Forouhar, F, Chinag, Y, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-11-25 | | Release date: | 2009-01-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Orthorombic crystal structure of serine phosphatase of rna polymerase ii ctd from fly drosofila melanogaster. northeast structural genomics consortium target fr253.
To be Published
|
|
3FMV
 
 | | Crystal structure of the serine phosphatase of RNA polymerase II CTD (SSU72 superfamily) from Drosophila melanogaster. Monoclinic crystal form. Northeast Structural Genomics Consortium target FR253. | | Descriptor: | Serine phosphatase of RNA polymerase II CTD | | Authors: | Kuzin, A.P, Chen, Y, Seetharaman, J, Forouhar, F, Chinag, Y, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-22 | | Release date: | 2009-01-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Crystal structure of the serine phosphatase of RNA polymerase II CTD (SSU72 superfamily) from Drosophila melanogaster. Monoclinic crystal form. Northeast Structural Genomics Consortium target FR253.
To be Published
|
|
1XQB
 
 | | X-Ray Structure Of YaeB from Haemophilus influenzae. Northeast Structural Genomics Research Consortium (NESGC)target IR47. | | Descriptor: | Hypothetical UPF0066 protein HI0510 | | Authors: | Benach, J, Lee, I, Forouhar, F, Kuzin, A.P, Keller, J.P, Itkin, A, Xiao, R, Acton, T, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-11 | | Release date: | 2004-10-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | X-Ray Structure Of YaeB from Haemophilus influenzae. Northeast Structural Genomics Research Consortium (NESGC) target IR47.
To be Published
|
|
