6JXR
 
 | | Structure of human T cell receptor-CD3 complex | | Descriptor: | T cell receptor alpha variable 12-3,Possible J 11 gene segment,T cell receptor alpha constant, T cell receptor beta variable 6-5,M1-specific T cell receptor beta chain,T cell receptor beta constant 2, T-cell surface glycoprotein CD3 delta chain, ... | | Authors: | Dong, D, Zheng, L, Lin, J, Zhu, Y, Li, N, Zhang, B, Xie, S, Zheng, J, Wang, Y, Gao, N, Huang, Z. | | Deposit date: | 2019-04-24 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of assembly of the human T cell receptor-CD3 complex.
Nature, 573, 2019
|
|
6K2U
 
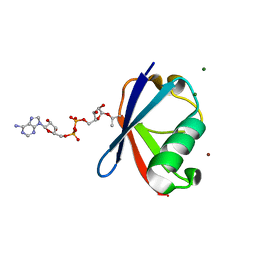 | | Crystal structure of Thr66 ADP-ribosylated ubiquitin | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, MAGNESIUM ION, Polyubiquitin-C, ... | | Authors: | Wang, X, Zhou, Y, Zhu, Y. | | Deposit date: | 2019-05-15 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.554 Å) | | Cite: | Threonine ADP-Ribosylation of Ubiquitin by a Bacterial Effector Family Blocks Host Ubiquitination.
Mol.Cell, 78, 2020
|
|
6JJ8
 
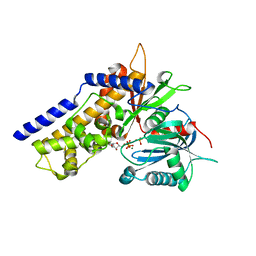 | | Crystal structure of OsHXK6-ATP-Mg2+ complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | He, C, Wei, P, Chen, J, Wang, H, Wan, Y, Zhou, J, Zhu, Y, Huang, W, Yin, L. | | Deposit date: | 2019-02-25 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of OsHXK6-ATP-Mg2+ complex
To Be Published
|
|
6JC3
 
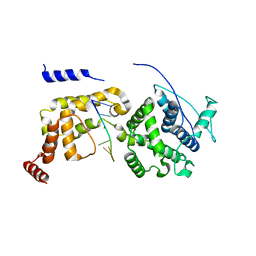 | | The Cryo-EM structure of nucleoprotein-RNA complex of Newcastle disease virus | | Descriptor: | Nucleocapsid, polyU | | Authors: | Song, X, Shan, H, Zhu, Y, Ding, W, Ouyang, S, Shen, Q.T, Liu, Z.J. | | Deposit date: | 2019-01-28 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Self-capping of nucleoprotein filaments protects the Newcastle disease virus genome.
Elife, 8, 2019
|
|
6JJ7
 
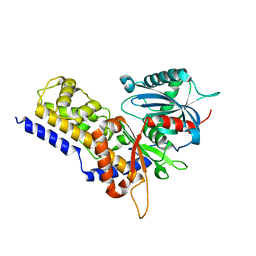 | | Crystal structure of OsHXK6-Glc complex | | Descriptor: | Rice hexokinase 6, beta-D-glucopyranose | | Authors: | He, C, Wei, P, Chen, J, Wang, H, Wan, Y, Zhou, J, Zhu, Y, Huang, W, Yin, L. | | Deposit date: | 2019-02-25 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of OsHXK6-Glc complex
To Be Published
|
|
8W72
 
 | |
8W6Z
 
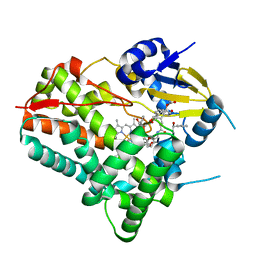 | | Substrate-bound crystal structure of a P450 enzyme DmlH that catalyze intramolecular phenol coupling in the biosynthesis of cihanmycins | | Descriptor: | (E)-3-[2-[(2R,3S)-3-[(1R)-1-aminocarbonyloxypropyl]oxiran-2-yl]phenyl]prop-2-enoic acid, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Fang, C, Zhang, L, Zhu, Y, Zhang, C. | | Deposit date: | 2023-08-30 | | Release date: | 2024-06-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate-bound crystal structure of a P450 enzyme DmlH that catalyze intramolecular phenol coupling in the biosynthesis of cihanmycins
To Be Published
|
|
8IAZ
 
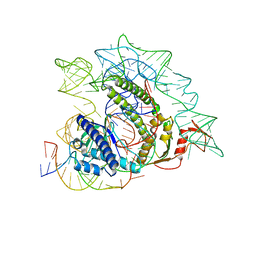 | | Cryo-EM structure of the ISFba1 TnpB-reRNA-dsDNA complex | | Descriptor: | DNA (5'-D(P*AP*CP*AP*TP*GP*GP*AP*CP*CP*AP*TP*CP*AP*GP*CP*TP*CP*CP*TP*AP*AP*TP*GP*G)-3'), DNA (5'-D(P*CP*CP*AP*TP*TP*AP*GP*GP*AP*GP*CP*TP*GP*AP*TP*G)-3'), RNA (207-MER), ... | | Authors: | Yin, M, Zhou, F, Zhu, Y, Huang, Z. | | Deposit date: | 2023-02-09 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Discovery and structural mechanism of DNA endonucleases guided by RAGATH-18-derived RNAs.
Cell Res., 34, 2024
|
|
5YEI
 
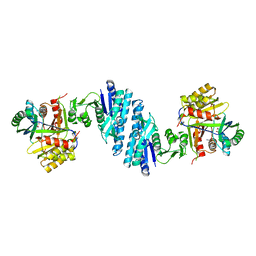 | | Mechanistic insight into the regulation of Pseudomonas aeruginosa aspartate kinase | | Descriptor: | Aspartokinase, GLYCEROL, LYSINE, ... | | Authors: | Li, C, Yang, M, Liu, L, Peng, C, Li, T, He, L, Song, Y, Zhu, Y, Bao, R. | | Deposit date: | 2017-09-17 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Mechanistic insights into the allosteric regulation of Pseudomonas aeruginosa aspartate kinase.
Biochem.J., 475, 2018
|
|
7W6K
 
 | | Cryo-EM structure of GmALMT12/QUAC1 anion channel | | Descriptor: | GmALMT12/QUAC1 | | Authors: | Qin, L, Tang, L.H, Xu, J.S, Zhang, X.H, Zhu, Y, Sun, F, Su, M, Zhai, Y.J, Chen, Y.H. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure and electrophysiological characterization of ALMT from Glycine max reveal a previously uncharacterized class of anion channels.
Sci Adv, 8, 2022
|
|
7WSO
 
 | | Structure of a membrane protein G | | Descriptor: | B-cell antigen receptor complex-associated protein alpha chain, B-cell antigen receptor complex-associated protein beta chain, Immunoglobulin heavy constant gamma 1 | | Authors: | Ma, X, Zhu, Y, Chen, Y, Huang, Z. | | Deposit date: | 2022-01-30 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Cryo-EM structures of two human B cell receptor isotypes.
Science, 377, 2022
|
|
7XBW
 
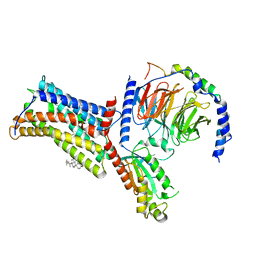 | | Cryo-EM structure of the human chemokine receptor CX3CR1 in complex with Gi1 | | Descriptor: | CHOLESTEROL, CX3C chemokine receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Lu, M, Zhao, W, Han, S, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2022-03-22 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Activation of the human chemokine receptor CX3CR1 regulated by cholesterol.
Sci Adv, 8, 2022
|
|
7XBX
 
 | | Cryo-EM structure of the human chemokine receptor CX3CR1 in complex with CX3CL1 and Gi1 | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Lu, M, Zhao, W, Han, S, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2022-03-22 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Activation of the human chemokine receptor CX3CR1 regulated by cholesterol.
Sci Adv, 8, 2022
|
|
5XNB
 
 | |
6IUF
 
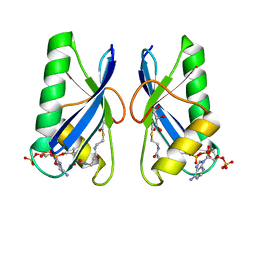 | | Crystal structure of Anti-CRISPR protein AcrVA5 | | Descriptor: | ACETYL COENZYME *A, GLYCEROL, protein-a | | Authors: | Dong, L, Guan, X, Zhu, Y, Huang, Z. | | Deposit date: | 2018-11-28 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | An anti-CRISPR protein disables type V Cas12a by acetylation.
Nat. Struct. Mol. Biol., 26, 2019
|
|
6IV6
 
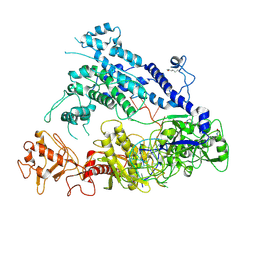 | | Cryo-EM structure of AcrVA5-acetylated MbCas12a in complex with crRNA | | Descriptor: | RNA (59-MER), nuclease | | Authors: | Dong, L, Li, N, Guan, X, Zhu, Y, Gao, N, Huang, Z. | | Deposit date: | 2018-12-02 | | Release date: | 2019-04-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | An anti-CRISPR protein disables type V Cas12a by acetylation.
Nat. Struct. Mol. Biol., 26, 2019
|
|
6AEG
 
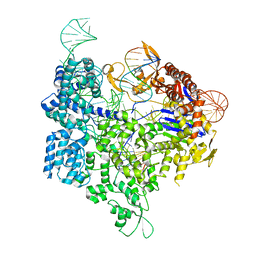 | | Crystal structure of xCas9 in complex with sgRNA and target DNA (GAT PAM) | | Descriptor: | DNA (25-MER), DNA (5'-D(*AP*AP*AP*GP*AP*TP*TP*AP*TP*TP*G)-3'), DNA nuclease, ... | | Authors: | Guo, M, Ren, K, Zhu, Y, Huang, Z. | | Deposit date: | 2018-08-04 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Structural insights into a high fidelity variant of SpCas9.
Cell Res., 29, 2019
|
|
6JEC
 
 | |
6JKL
 
 | |
6AEB
 
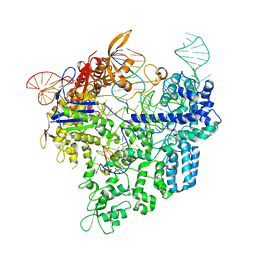 | | Crystal structure of xCas9 in complex with sgRNA and target DNA (AAG PAM) | | Descriptor: | DNA (25-MER), DNA (5'-D(*AP*AP*AP*AP*AP*GP*TP*AP*TP*TP*G)-3'), DNA Nuclease, ... | | Authors: | Guo, M, Ren, K, Zhu, Y, Huang, Z. | | Deposit date: | 2018-08-04 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | Structural insights into a high fidelity variant of SpCas9.
Cell Res., 29, 2019
|
|
5B7I
 
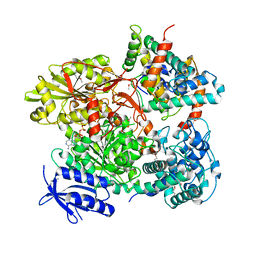 | | Cas3-AcrF3 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, CRISPR-associated nuclease/helicase Cas3 subtype I-F/YPEST, ... | | Authors: | Wang, X, Zhu, Y. | | Deposit date: | 2016-06-07 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of Cas3 inhibition by the bacteriophage protein AcrF3
Nat.Struct.Mol.Biol., 23, 2016
|
|
7VOP
 
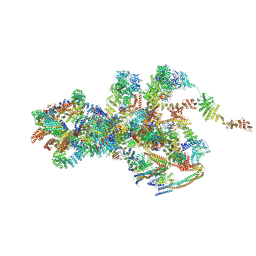 | | Cryo-EM structure of Xenopus laevis nuclear pore complex cytoplasmic ring subunit | | Descriptor: | GATOR complex protein SEC13, IL4I1 protein, MGC154553 protein, ... | | Authors: | Tai, L, Zhu, Y, Sun, F. | | Deposit date: | 2021-10-14 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | 8 angstrom structure of the outer rings of the Xenopus laevis nuclear pore complex obtained by cryo-EM and AI.
Protein Cell, 13, 2022
|
|
7VCI
 
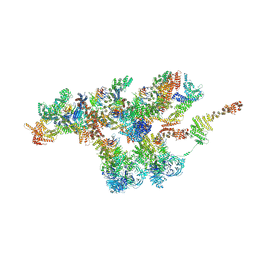 | | Structure of Xenopus laevis NPC nuclear ring asymmetric unit | | Descriptor: | GATOR complex protein SEC13, MGC154553 protein, MGC83295 protein, ... | | Authors: | Tai, L, Zhu, Y, Sun, F. | | Deposit date: | 2021-09-03 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | 8 angstrom structure of the outer rings of the Xenopus laevis nuclear pore complex obtained by cryo-EM and AI.
Protein Cell, 13, 2022
|
|
7FJD
 
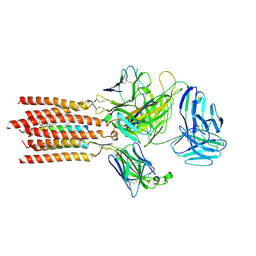 | | Cryo-EM structure of a membrane protein(WT) | | Descriptor: | CHOLESTEROL, T cell receptor alpha variable 12-3,Possible J 11 gene segment,T cell receptor alpha chain constant, T cell receptor beta variable 6-5,M1-specific T cell receptor beta chain,T cell receptor beta constant 2, ... | | Authors: | Chen, Y, Zhu, Y, Gao, W, Zhang, A, Guo, C, Huang, Z. | | Deposit date: | 2021-08-03 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cholesterol inhibits TCR signaling by directly restricting TCR-CD3 core tunnel motility.
Mol.Cell, 82, 2022
|
|
7FJE
 
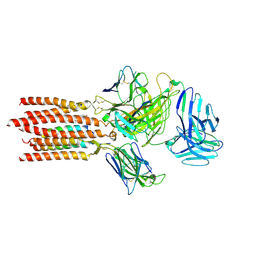 | | Cryo-EM structure of a membrane protein(LL) | | Descriptor: | CHOLESTEROL, T cell receptor alpha variable 12-3,Possible J 11 gene segment,T cell receptor alpha chain constant, T cell receptor beta variable 6-5,M1-specific T cell receptor beta chain,T cell receptor beta constant 2, ... | | Authors: | Chen, Y, Zhu, Y, Gao, W, Zhang, A, Guo, C, Huang, Z. | | Deposit date: | 2021-08-03 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cholesterol inhibits TCR signaling by directly restricting TCR-CD3 core tunnel motility.
Mol.Cell, 82, 2022
|
|
