1MDY
 
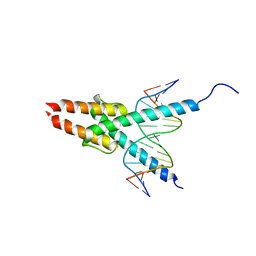 | | CRYSTAL STRUCTURE OF MYOD BHLH DOMAIN BOUND TO DNA: PERSPECTIVES ON DNA RECOGNITION AND IMPLICATIONS FOR TRANSCRIPTIONAL ACTIVATION | | Descriptor: | DNA (5'-D(*TP*CP*AP*AP*CP*AP*GP*CP*TP*GP*TP*TP*GP*A)-3'), PROTEIN (MYOD BHLH DOMAIN) | | Authors: | Ma, P.C.M, Rould, M.A, Weintraub, H, Pabo, C.O. | | Deposit date: | 1994-06-09 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of MyoD bHLH domain-DNA complex: perspectives on DNA recognition and implications for transcriptional activation.
Cell(Cambridge,Mass.), 77, 1994
|
|
1HLO
 
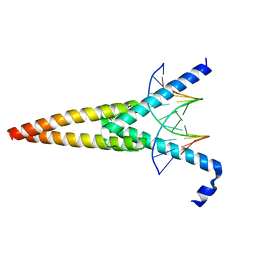 | | THE CRYSTAL STRUCTURE OF AN INTACT HUMAN MAX-DNA COMPLEX: NEW INSIGHTS INTO MECHANISMS OF TRANSCRIPTIONAL CONTROL | | Descriptor: | DNA (5'-D(*AP*CP*CP*AP*CP*GP*TP*GP*GP*TP*G)-3'), DNA (5'-D(*CP*AP*CP*CP*AP*CP*GP*TP*GP*GP*T)-3'), PROTEIN (TRANSCRIPTION FACTOR MAX) | | Authors: | Brownlie, P, Ceska, T.A, Lamers, M, Romier, C, Theo, H, Suck, D. | | Deposit date: | 1997-09-10 | | Release date: | 1997-10-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of an intact human Max-DNA complex: new insights into mechanisms of transcriptional control.
Structure, 5, 1997
|
|
1A0A
 
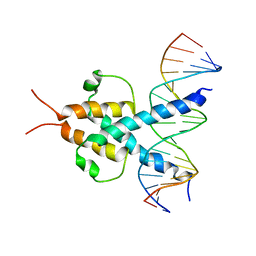 | | PHOSPHATE SYSTEM POSITIVE REGULATORY PROTEIN PHO4/DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*TP*AP*GP*TP*CP*CP*CP*AP*CP*GP*TP*GP*TP*GP*AP*G )-3'), DNA (5'-D(*CP*TP*CP*AP*CP*AP*CP*GP*TP*GP*GP*GP*AP*CP*TP*AP*G )-3'), PROTEIN (PHOSPHATE SYSTEM POSITIVE REGULATORY PROTEIN PHO4) | | Authors: | Shimizu, T, Toumoto, A, Ihara, K, Shimizu, M, Kyogoku, Y, Ogawa, N, Oshima, Y, Hakoshima, T. | | Deposit date: | 1997-11-27 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of PHO4 bHLH domain-DNA complex: flanking base recognition.
EMBO J., 16, 1997
|
|
1NKP
 
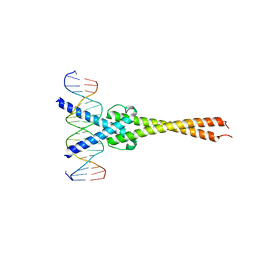 | | Crystal structure of Myc-Max recognizing DNA | | Descriptor: | 5'-D(*CP*GP*AP*GP*TP*AP*GP*CP*AP*CP*GP*TP*GP*CP*TP*AP*CP*TP*C)-3', Max protein, Myc proto-oncogene protein | | Authors: | Nair, S.K, Burley, S.K. | | Deposit date: | 2003-01-03 | | Release date: | 2003-02-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structures of Myc-Max and Mad-Max recognizing DNA: Molecular bases of regulation by proto-oncogenic transcription factors
Cell(Cambridge,Mass.), 112, 2003
|
|
1NLW
 
 | | Crystal structure of Mad-Max recognizing DNA | | Descriptor: | 5'-D(*GP*AP*GP*TP*AP*GP*CP*AP*CP*GP*TP*GP*CP*TP*AP*CP*TP*C)-3', MAD PROTEIN, MAX PROTEIN | | Authors: | Nair, S.K, Burley, S.K. | | Deposit date: | 2003-01-07 | | Release date: | 2003-02-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structures of Myc-Max and Mad-Max recognizing DNA: Molecular bases of regulation by proto-oncogenic transcription factors
Cell(Cambridge,Mass.), 112, 2003
|
|
1X0O
 
 | | human ARNT C-terminal PAS domain | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator | | Authors: | Card, P.B, Erbel, P.J, Gardner, K.H. | | Deposit date: | 2005-03-25 | | Release date: | 2005-10-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of ARNT PAS-B Dimerization: Use of a Common Beta-sheet Interface for Hetero- and Homodimerization.
J.Mol.Biol., 353, 2005
|
|
2A24
 
 | | HADDOCK Structure of HIF-2a/ARNT PAS-B Heterodimer | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain protein 1 | | Authors: | Card, P.B, Erbel, P.J, Gardner, K.H. | | Deposit date: | 2005-06-21 | | Release date: | 2006-01-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of ARNT PAS-B dimerization: use of a common beta-sheet interface for hetero- and homodimerization.
J.Mol.Biol., 353, 2005
|
|
2QL2
 
 | | Crystal Structure of the basic-helix-loop-helix domains of the heterodimer E47/NeuroD1 bound to DNA | | Descriptor: | DNA (5'-D(*DAP*DGP*DGP*DAP*DCP*DCP*DAP*DGP*DAP*DTP*DGP*DGP*DCP*DCP*DTP*DA)-3'), DNA (5'-D(*DTP*DAP*DGP*DGP*DCP*DCP*DAP*DTP*DCP*DTP*DGP*DGP*DTP*DCP*DCP*DT)-3'), Neurogenic differentiation factor 1, ... | | Authors: | Rose, R.B, Longo, A, Guanga, G.P. | | Deposit date: | 2007-07-12 | | Release date: | 2008-11-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of E47-NeuroD1/beta2 bHLH domain-DNA complex: heterodimer selectivity and DNA recognition.
Biochemistry, 47, 2008
|
|
2K7S
 
 | |
3F1O
 
 | | Crystal structure of the high affinity heterodimer of HIF2 alpha and ARNT C-terminal PAS domains, with an internally-bound artificial ligand | | Descriptor: | 1,2-ETHANEDIOL, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1, ... | | Authors: | Scheuermann, T.H, Tomchick, D.R, Machius, M, Guo, Y, Bruick, R.K, Gardner, K.H. | | Deposit date: | 2008-10-28 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Artificial ligand binding within the HIF2alpha PAS-B domain of the HIF2 transcription factor.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3F1N
 
 | | Crystal structure of a high affinity heterodimer of HIF2 alpha and ARNT C-terminal PAS domains, with internally bound ethylene glycol. | | Descriptor: | 1,2-ETHANEDIOL, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Scheuermann, T.H, Tomchick, D.R, Machius, M, Guo, Y, Bruick, R.K, Gardner, K.H. | | Deposit date: | 2008-10-28 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.479 Å) | | Cite: | Artificial ligand binding within the HIF2alpha PAS-B domain of the HIF2 transcription factor.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3F1P
 
 | | Crystal structure of a high affinity heterodimer of HIF2 alpha and ARNT C-terminal PAS domains | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Scheuermann, T.H, Tomchick, D.R, Machius, M, Guo, Y, Bruick, R.K, Gardner, K.H. | | Deposit date: | 2008-10-28 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Artificial ligand binding within the HIF2alpha PAS-B domain of the HIF2 transcription factor.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3H7W
 
 | | Crystal structure of the high affinity heterodimer of HIF2 alpha and ARNT C-terminal PAS domains with the artificial ligand THS017 | | Descriptor: | 2-nitro-N-(thiophen-3-ylmethyl)-4-(trifluoromethyl)aniline, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Key, J.M, Scheuermann, T.H, Anderson, P.C, Daggett, V, Gardner, K.H. | | Deposit date: | 2009-04-28 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Principles of ligand binding within a completely buried cavity in HIF2alpha PAS-B
J.Am.Chem.Soc., 131, 2009
|
|
3H82
 
 | | Crystal structure of the high affinity heterodimer of HIF2 alpha and ARNT C-terminal PAS domains with the artificial ligand THS020 | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1, N-(furan-2-ylmethyl)-2-nitro-4-(trifluoromethyl)aniline | | Authors: | Key, J.M, Scheuermann, T.H, Anderson, P.C, Daggett, V, Gardner, K.H. | | Deposit date: | 2009-04-28 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Principles of ligand binding within a completely buried cavity in HIF2alpha PAS-B
J.Am.Chem.Soc., 131, 2009
|
|
3L03
 
 | | Crystal Structure of human Estrogen Receptor alpha Ligand-Binding Domain in complex with a Glucocorticoid Receptor Interacting Protein 1 Nr Box II peptide and Estetrol (Estra-1,3,5(10)-triene-3,15 alpha,16alpha,17beta-tetrol) | | Descriptor: | (14beta,15alpha,16alpha,17alpha)-estra-1,3,5(10)-triene-3,15,16,17-tetrol, CHLORIDE ION, Estrogen receptor, ... | | Authors: | Rajan, S.S, Kim, Y, Vanek, K, Joachimiak, A, Greene, G.L. | | Deposit date: | 2009-12-09 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Crystal Structure of human Estrogen Receptor alpha Ligand-Binding Domain
in complex with a Glucocorticoid Receptor Interacting Protein 1 Nr Box II
peptide and Estra-1,3,5(10)-triene-3,15 alpha,16alpha,17beta-tetrol
To be Published
|
|
3LMP
 
 | | Crystal structure of the PPARgamma-LBD complexed with a cercosporamide derivative modulator | | Descriptor: | (9aS)-8-acetyl-1,7-dihydroxy-3-methoxy-9a-methyl-N-(1-naphthylmethyl)-9-oxo-9,9a-dihydrodibenzo[b,d]furan-4-carboxamide, Peptide of Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Matsui, Y, Hanzawa, H. | | Deposit date: | 2010-01-31 | | Release date: | 2010-04-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a novel selective PPARgamma modulator from (-)-Cercosporamide derivatives
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2KWF
 
 | | The structure of E-protein activation domain 1 bound to the KIX domain of CBP/p300 elucidates leukemia induction by E2A-PBX1 | | Descriptor: | CREB-binding protein, Transcription factor 4 | | Authors: | Denis, C.M, Chitayat, S, Plevin, M.J, Liu, S, Spencer, H.L, Ikura, M, LeBrun, D.P, Smith, S.P. | | Deposit date: | 2010-04-08 | | Release date: | 2011-07-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The structure of E-protein activation domain 1 bound to the KIX domain of CBP/p300 elucidates leukemia induction by E2A-PBX1
To be Published
|
|
3O1D
 
 | | Structure-function study of Gemini derivatives with two different side chains at C-20, Gemini-0072 and Gemini-0097. | | Descriptor: | (1R,3R,7E,17beta)-17-[(1S)-6,6,6-trifluoro-5-hydroxy-1-(4-hydroxy-4-methylpentyl)-5-(trifluoromethyl)hex-3-yn-1-yl]-9,10-secoestra-5,7-diene-1,3-diol, Nuclear receptor coactivator 2, Vitamin D3 receptor A | | Authors: | Huet, T, Moras, D, Rochel, N. | | Deposit date: | 2010-07-21 | | Release date: | 2011-07-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4007 Å) | | Cite: | Structure-function study of gemini derivatives with two different side chains at C-20, Gemini-0072 and Gemini-0097.
Medchemcomm, 2, 2011
|
|
3O1E
 
 | | Structure-function of Gemini derivatives with two different side chains at C-20, Gemini-0072 and Gemini-0097. | | Descriptor: | (1R,3R,7E,17beta)-17-[(1R)-6,6,6-trifluoro-5-hydroxy-1-(4-hydroxy-4-methylpentyl)-5-(trifluoromethyl)hex-3-yn-1-yl]-9,1 0-secoestra-5,7-diene-1,3-diol, Nuclear receptor coactivator 2, Vitamin D3 receptor A | | Authors: | Huet, T, Moras, D, Rochel, N. | | Deposit date: | 2010-07-21 | | Release date: | 2011-07-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5001 Å) | | Cite: | Structure-function study of gemini derivatives with two different side chains at C-20, Gemini-0072 and Gemini-0097.
Medchemcomm, 2, 2011
|
|
3OAP
 
 | | Crystal structure of human Retinoid X Receptor alpha-ligand binding domain complex with 9-cis retinoic acid and the coactivator peptide GRIP-1 | | Descriptor: | (9cis)-retinoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Xia, G, Smith, C.D, Muccio, D.D. | | Deposit date: | 2010-08-05 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure, Energetics and Dynamics of Binding Coactivator Peptide to Human Retinoid X Receptor Alpha Ligand Binding Domain Complex with 9-cis-Retinoic Acid.
Biochemistry, 50, 2011
|
|
3OKI
 
 | |
3OKH
 
 | |
3OLL
 
 | | Crystal structure of phosphorylated estrogen receptor beta ligand binding domain | | Descriptor: | ESTRADIOL, Estrogen receptor beta, Nuclear receptor coactivator 1 | | Authors: | Moecklinghoff, S, Rose, R, Ottmann, C, Brunsveld, L. | | Deposit date: | 2010-08-26 | | Release date: | 2010-11-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Synthesis and crystal structure of a phosphorylated estrogen receptor ligand binding domain.
Chembiochem, 11, 2010
|
|
3OLF
 
 | |
3OLS
 
 | | Crystal structure of estrogen receptor beta ligand binding domain | | Descriptor: | ESTRADIOL, Estrogen receptor beta, Nuclear receptor coactivator 1 | | Authors: | Moecklinghoff, S, Rose, R, Carraz, M, Visser, A, Ottmann, C, Brunsveld, L. | | Deposit date: | 2010-08-26 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis and crystal structure of a phosphorylated estrogen receptor ligand binding domain.
Chembiochem, 11, 2010
|
|
