9G7H
 
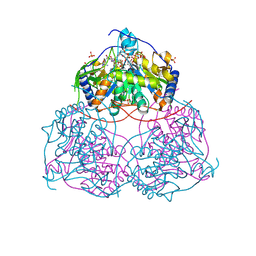 | | Human Sirt6 in complex with ADP-ribose and the inhibitor 2-Pr | | Descriptor: | 2,4-bis(oxidanylidene)-1-[2-oxidanylidene-2-[[(2S)-3-oxidanylidene-3-(propylamino)-2-[3-(3-pyridin-3-yl-1,2,4-oxadiazol-5-yl)propanoylamino]propyl]amino]ethyl]pyrimidine-5-carboxamide, NAD-dependent protein deacetylase sirtuin-6, SULFATE ION, ... | | Authors: | You, W, Steegborn, C. | | Deposit date: | 2024-07-21 | | Release date: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Elucidating the Unconventional Binding Mode of a DNA-Encoded Library Hit Provides a Blueprint for Sirtuin 6 Inhibitor Development.
Chemmedchem, 2024
|
|
9CBT
 
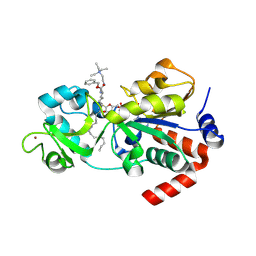 | | Crystal structure of human sirtuin 3 fragment (residues 118-399) bound to intermediates from reaction with NAD and inhibitor NH6-10 | | Descriptor: | (phenylmethyl) ~{N}-[(2~{S})-6-[[(2~{R},3~{a}~{R},5~{R},6~{R},6~{a}~{R})-5-[[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-6-oxidanyl-2-tridecyl-3~{a},5,6,6~{a}-tetrahydrofuro[2,3-d][1,3]oxathiol-2-yl]amino]-1-oxidanylidene-1-[2-(triethyl-$l^{4}-azanyl)ethylamino]hexan-2-yl]carbamate, 2-{[(2S)-6-[(Z)-(1-{[(2R,3R,4R,5R)-5-({[(R)-{[(R)-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}methyl)-4-hydroxy-2-sulfanyloxolan-3-yl]oxy}tetradecylidene)amino]-2-{[(benzyloxy)carbonyl]amino}hexanoyl]amino}-N,N,N-triethylethan-1-aminium (non-preferred name), NAD-dependent protein deacetylase sirtuin-3, ... | | Authors: | Fenwick, M.K, Young, H.J, Lin, H. | | Deposit date: | 2024-06-20 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of human sirtuin 3 fragment (residues 118-399) bound to intermediates from reaction with NAD and inhibitor NH6-10
To Be Published
|
|
8XE7
 
 | |
8V2N
 
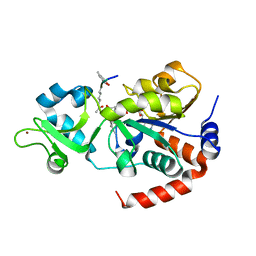 | | Human SIRT3 co-crystallized with ligands, including p53-AMC peptide and Carba-NAD | | Descriptor: | CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, GLN-PRO-LYS-FDL, NAD-dependent protein deacetylase sirtuin-3, ... | | Authors: | Chakrabarti, R, Ghosh, A, Guan, X, Upadhyay, A, Dumpati, R.K, Munshi, S, Roy, S, Chall, S, Rahnamoun, A, Reverdy, C, Errasti, G, Delacroix, T. | | Deposit date: | 2023-11-23 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Computationally Driven Discovery and Characterization of SIRT3 Activating Compounds that Fully Recover Catalytic Activity under NAD+ Depletion
biorxiv, 2023
|
|
8V15
 
 | | Human SIRT3 bound to p53-AMC peptide, Carba-NAD, and Honokiol | | Descriptor: | (1P)-3',5-di(prop-2-en-1-yl)[1,1'-biphenyl]-2,4'-diol, CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, GLN-PRO-LYS-FDL, ... | | Authors: | Chakrabarti, R, Ghosh, A, Guan, X, Upadhyay, A, Dumpati, R.K, Munshi, S, Roy, S, Chall, S, Rahnamoun, A, Reverdy, C, Errasti, G, Delacroix, T. | | Deposit date: | 2023-11-19 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Computationally Driven Discovery and Characterization of SIRT3 Activating Compounds that Fully Recover Catalytic Activity under NAD+ Depletion
biorxiv, 2023
|
|
8TGP
 
 | |
8PY3
 
 | | Crystal structure of human Sirt2 in complex with a 1,2,4-oxadiazole based inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-chloranyl-~{N}-[4-[5-[[(3~{S})-1-[(3-fluoranyl-2-methyl-phenyl)methyl]piperidin-3-yl]methyl]-1,2,4-oxadiazol-3-yl]phenyl]benzamide, ... | | Authors: | Friedrich, F, Colcerasa, A, Einsle, O, Jung, M. | | Deposit date: | 2023-07-24 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Activity Studies of 1,2,4-Oxadiazoles for the Inhibition of the NAD + -Dependent Lysine Deacylase Sirtuin 2.
J.Med.Chem., 67, 2024
|
|
8OWZ
 
 | | Crystal structure of human Sirt2 in complex with a triazole-based SirReal | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 1,2-ETHANEDIOL, 2-(4,6-dimethylpyrimidin-2-yl)sulfanyl-N-[5-[[3-[[1-(2-methoxyethyl)-1,2,3-triazol-4-yl]methoxy]phenyl]methyl]-1,3-thiazol-2-yl]ethanamide, ... | | Authors: | Friedrich, F, Zhang, L, Schiedel, M, Einsle, O, Jung, M. | | Deposit date: | 2023-04-28 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Development of First-in-Class Dual Sirt2/HDAC6 Inhibitors as Molecular Tools for Dual Inhibition of Tubulin Deacetylation.
J.Med.Chem., 66, 2023
|
|
8OF4
 
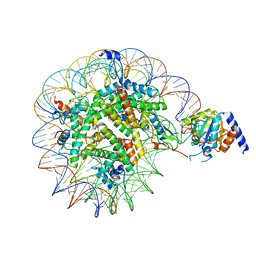 | | Nucleosome Bound human SIRT6 (Composite) | | Descriptor: | DNA (145-MER), Histone H2A type 1, Histone H2B, ... | | Authors: | Smirnova, E, Bignon, E, Schultz, P, Papai, G, Ben-Shem, A. | | Deposit date: | 2023-03-13 | | Release date: | 2023-08-09 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Binding to nucleosome poises human SIRT6 for histone H3 deacetylation.
Elife, 12, 2024
|
|
8I2B
 
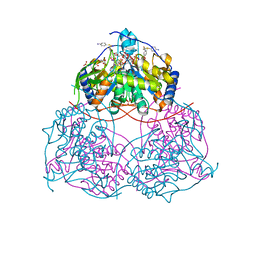 | | Human SIRT6 in complex with inhibitor 7702 | | Descriptor: | N1-[[4-(4-aminophenyl)sulfanyl-3-(trifluoromethyl)phenyl]methoxy]benzene-1,4-dicarboxamide, NAD-dependent protein deacylase sirtuin-6, SULFATE ION, ... | | Authors: | Wang, Y. | | Deposit date: | 2023-01-14 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The SIRT6 in complex with inhibitor 7702
To Be Published
|
|
8HN9
 
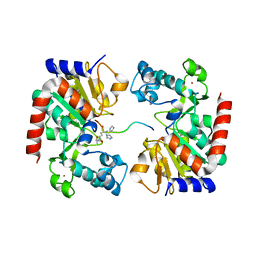 | | Human SIRT3 Recognizing CCNE2K348la peptide | | Descriptor: | CCNE2 peptide, IMIDAZOLE, NAD-dependent protein deacetylase sirtuin-3, ... | | Authors: | Wang, Y, Ding, W. | | Deposit date: | 2022-12-07 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | SIRT3-dependent delactylation of cyclin E2 prevents hepatocellular carcinoma growth.
Embo Rep., 24, 2023
|
|
8HLY
 
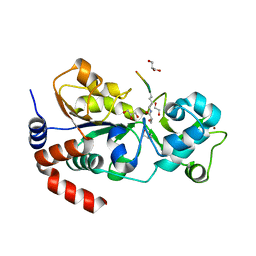 | |
8HLW
 
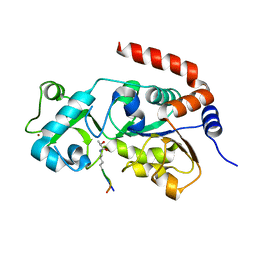 | | Crystal structure of SIRT3 in complex with H4K16la peptide | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, Histone H4 residues 20-27, NAD-dependent protein deacetylase sirtuin-3, ... | | Authors: | Zhuming, F, Hao, Q. | | Deposit date: | 2022-12-01 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of SIRT3 as an eraser of H4K16la.
Iscience, 26, 2023
|
|
8GBN
 
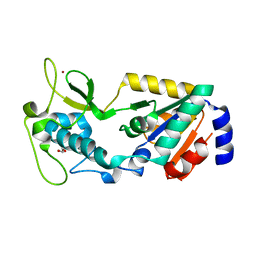 | | Structure of Apo Human SIRT5 P114T Mutant | | Descriptor: | 1,2-ETHANEDIOL, NAD-dependent protein deacylase sirtuin-5, mitochondrial, ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2023-02-26 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human SIRT5 variants with reduced stability and activity do not cause neuropathology in mice.
Iscience, 27, 2024
|
|
8GBL
 
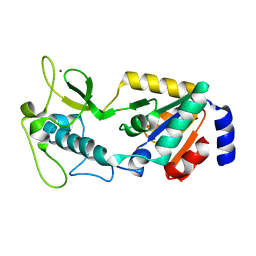 | | Structure of Apo Human SIRT5 | | Descriptor: | NAD-dependent protein deacylase sirtuin-5, mitochondrial, ZINC ION | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2023-02-26 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Human SIRT5 variants with reduced stability and activity do not cause neuropathology in mice.
Iscience, 27, 2024
|
|
8G57
 
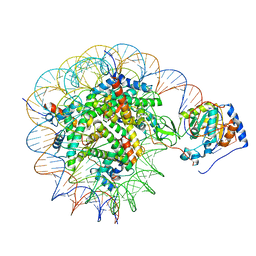 | | Structure of nucleosome-bound Sirtuin 6 deacetylase | | Descriptor: | DNA strand 1, DNA strand 2, Histone H2A type 1-B/E, ... | | Authors: | Chio, U.S, Rechiche, O, Bryll, A.R, Zhu, J, Feldman, J.L, Peterson, C.L, Tan, S, Armache, J.-P. | | Deposit date: | 2023-02-11 | | Release date: | 2023-04-26 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Cryo-EM structure of the human Sirtuin 6-nucleosome complex.
Sci Adv, 9, 2023
|
|
8F86
 
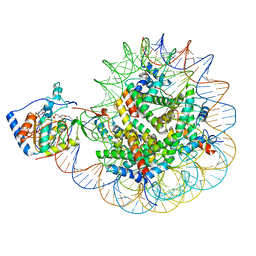 | | SIRT6 bound to an H3K9Ac nucleosome | | Descriptor: | DNA (148-MER), Histone H2A type 1, Histone H2B, ... | | Authors: | Markert, J, Whedon, S, Wang, Z, Cole, P, Farnung, L. | | Deposit date: | 2022-11-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Basis of Sirtuin 6-Catalyzed Nucleosome Deacetylation.
J.Am.Chem.Soc., 145, 2023
|
|
8CNO
 
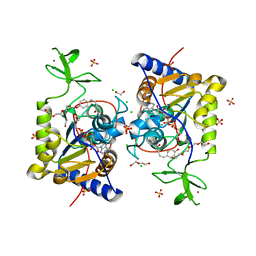 | |
8CNM
 
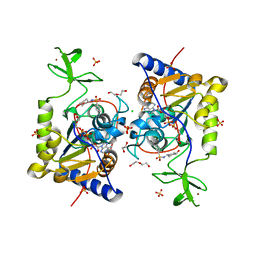 | |
8CCZ
 
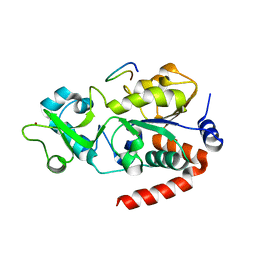 | |
8CCW
 
 | |
8BL1
 
 | | Human Sirt6 in complex with the inhibitor S6039 and ADP-ribose | | Descriptor: | 2-azanyl-5-[4-[[5-(2-fluorophenyl)thiophen-2-yl]methyl]piperazin-1-yl]sulfonyl-benzenesulfonamide, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | You, W, Steegborn, C. | | Deposit date: | 2022-11-09 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Development of novel Sirtuin 6 inhibitors and activators based on a protein crystallography-based fragment screen
To Be Published
|
|
8BL0
 
 | | Human Sirt6 in complex with the inhibitor S6023 and ADP-ribose | | Descriptor: | 3-[4-[[5-(2-fluorophenyl)thiophen-2-yl]methyl]piperazin-1-yl]sulfonylbenzenesulfonamide, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | You, W, Steegborn, C. | | Deposit date: | 2022-11-09 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Development of novel Sirtuin 6 inhibitors and activators based on a protein crystallography-based fragment screen
To Be Published
|
|
8BBK
 
 | |
8AKG
 
 | |
