1HQK
 
 | | CRYSTAL STRUCTURE ANALYSIS OF LUMAZINE SYNTHASE FROM AQUIFEX AEOLICUS | | Descriptor: | 6,7-DIMETHYL-8-RIBITYLLUMAZINE SYNTHASE | | Authors: | Zhang, X, Meining, W, Fischer, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2000-12-18 | | Release date: | 2001-12-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray structure analysis and crystallographic refinement of lumazine synthase from the hyperthermophile Aquifex aeolicus at 1.6 A resolution: determinants of thermostability revealed from structural comparisons.
J.Mol.Biol., 306, 2001
|
|
2C92
 
 | | LUMAZINE SYNTHASE FROM MYCOBACTERIUM TUBERCULOSIS BOUND TO 3-(1,3,7- TRIHYDRO-9-D-RIBITYL-2,6,8-PURINETRIONE-7-YL) PENTANE 1 PHOSPHATE | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 3-(1,3,7-TRIHYDRO-9-D-RIBITYL-2,6,8-PURINETRIONE-7-YL) 1-PHOSPHATE, ... | | Authors: | Morgunova, E, Illarionov, B, Jin, G, Haase, I, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2005-12-09 | | Release date: | 2006-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Thermodynamic Insights Into the Binding Mode of Five Novel Inhibitors of Lumazine Synthase from Mycobacterium Tuberculosis.
FEBS J., 273, 2006
|
|
1NQU
 
 | | Crystal Structure of Lumazine Synthase from Aquifex aeolicus in Complex with Inhibitor: 6,7-dioxo-5H-8-ribitylaminolumazine | | Descriptor: | 6,7-DIOXO-5H-8-RIBITYLAMINOLUMAZINE, 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Zhang, X, Meining, W, Cushman, M, Haase, I, Fischer, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2003-01-23 | | Release date: | 2004-01-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A structure-based model of the reaction catalyzed by lumazine synthase from Aquifex aeolicus.
J.Mol.Biol., 328, 2003
|
|
1NQX
 
 | | Crystal Structure of Lumazine Synthase from Aquifex aeolicus in Complex with Inhibitor: 3-(7-hydroxy-8-ribityllumazine-6-yl)propionic acid | | Descriptor: | 3-(7-HYDROXY-8-RIBITYLLUMAZINE-6-YL) PROPIONIC ACID, 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Zhang, X, Meining, W, Cushman, M, Haase, I, Fischer, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2003-01-23 | | Release date: | 2004-01-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A structure-based model of the reaction catalyzed by lumazine synthase from Aquifex aeolicus.
J.Mol.Biol., 328, 2003
|
|
1EJB
 
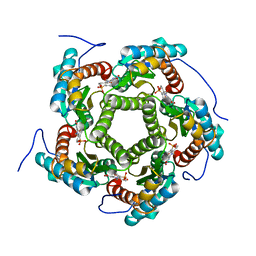 | | LUMAZINE SYNTHASE FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | 5-(6-D-RIBITYLAMINO-2,4-DIHYDROXYPYRIMIDIN-5-YL)-1-PENTYL-PHOSPHONIC ACID, LUMAZINE SYNTHASE | | Authors: | Meining, W, Mortl, S, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2000-03-02 | | Release date: | 2001-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The atomic structure of pentameric lumazine synthase from Saccharomyces cerevisiae at 1.85 A resolution reveals the binding mode of a phosphonate intermediate analogue.
J.Mol.Biol., 299, 2000
|
|
2C94
 
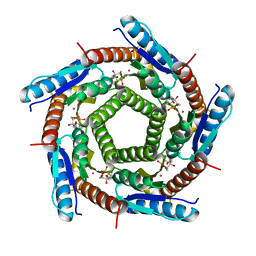 | | LUMAZINE SYNTHASE FROM MYCOBACTERIUM TUBERCULOSIS BOUND TO 3-(1,3,7- TRIHYDRO-9-D-RIBITYL-2,6,8-PURINETRIONE-7-YL) 1,1 difluoropentane-1- PHOSPHATE | | Descriptor: | 3-(1,3,7-TRIHYDRO-9-D-RIBITYL-2,6,8-PURINETRIONE-7-YL) 1,1 DIFLUOROPENTANE-1-PHOSPHATE, 6,7-DIMETHYL-8-RIBITYLLUMAZINE SYNTHASE, POTASSIUM ION | | Authors: | Morgunova, E, Illarionov, B, Jin, G, Haase, I, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2005-12-09 | | Release date: | 2006-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Thermodynamic Insights Into the Binding Mode of Five Novel Inhibitors of Lumazine Synthase from Mycobacterium Tuberculosis.
FEBS J., 273, 2006
|
|
4J07
 
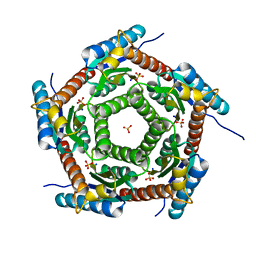 | | Crystal structure of a PROBABLE RIBOFLAVIN SYNTHASE, BETA CHAIN RIBH (6,7-dimethyl-8-ribityllumazine synthase, DMRL synthase, Lumazine synthase) from Mycobacterium leprae | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase, SODIUM ION, SULFATE ION | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2013-01-30 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of a Probable Riboflavin Synthase, Beta chain RIBH (6,7-dimethyl-8-ribityllumazine synthase, DMRL synthase, Lumazine synthase) from Mycobacterium leprae
TO BE PUBLISHED
|
|
1W19
 
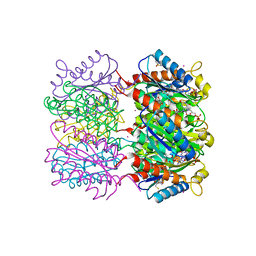 | | Lumazine Synthase from Mycobacterium tuberculosis bound to 3-(1,3,7- trihydro-9-D-ribityl-2,6,8-purinetrione-7-yl)propane 1-phosphate | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, (4S,5S)-1,2-DITHIANE-4,5-DIOL, ... | | Authors: | Morgunova, E, Meining, W, Illarionov, B, Haase, I, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2004-06-03 | | Release date: | 2005-03-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Lumazine Synthase from Mycobacterium Tuberculosis as a Target for Rational Drug Design: Binding Mode of a New Class of Purinetrione Inhibitors(,)
Biochemistry, 44, 2005
|
|
1KZ1
 
 | | Mutant enzyme W27G Lumazine Synthase from S.pombe | | Descriptor: | 6,7-Dimethyl-8-ribityllumazine Synthase | | Authors: | Gerhardt, S, Haase, I, Steinbacher, S, Kaiser, J.T, Cushman, M, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2002-02-06 | | Release date: | 2002-07-24 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural basis of riboflavin binding to Schizosaccharomyces pombe 6,7-dimethyl-8-ribityllumazine synthase.
J.Mol.Biol., 318, 2002
|
|
2C97
 
 | | LUMAZINE SYNTHASE FROM MYCOBACTERIUM TUBERCULOSIS BOUND TO 4-(6- chloro-2,4-dioxo-1,2,3,4-tetrahydropyrimidin-5-yl)butyl phosphate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(6-CHLORO-2,4-DIOXO-1,2,3,4-TETRAHYDROPYRIMIDIN-5-YL) BUTYL PHOSPHATE, 6,7-DIMETHYL-8-RIBITYLLUMAZINE SYNTHASE, ... | | Authors: | Morgunova, E, Illarionov, B, Jin, G, Haase, I, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2005-12-09 | | Release date: | 2006-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Thermodynamic Insights Into the Binding Mode of Five Novel Inhibitors of Lumazine Synthase from Mycobacterium Tuberculosis.
FEBS J., 273, 2006
|
|
1NQV
 
 | | Crystal Structure of Lumazine Synthase from Aquifex aeolicus in Complex with Inhibitor: 5-nitroso-6-ribityl-amino-2,4(1H,3H)pyrimidinedione | | Descriptor: | 5-NITROSO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Zhang, X, Meining, W, Cushman, M, Haase, I, Fischer, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2003-01-23 | | Release date: | 2004-01-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A structure-based model of the reaction catalyzed by lumazine synthase from Aquifex aeolicus.
J.Mol.Biol., 328, 2003
|
|
1NQW
 
 | | Crystal Structure of Lumazine Synthase from Aquifex aeolicus in Complex with Inhibitor: 5-(6-D-ribitylamino-2,4(1H,3H)pyrimidinedione-5-yl)-1-pentyl-phosphonic acid | | Descriptor: | 5-(6-D-RIBITYLAMINO-2,4(1H,3H)PYRIMIDINEDIONE-5-YL) PENTYL-1-PHOSPHONIC ACID, 6,7-dimethyl-8-ribityllumazine synthase | | Authors: | Zhang, X, Meining, W, Cushman, M, Haase, I, Fischer, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2003-01-23 | | Release date: | 2004-01-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A structure-based model of the reaction catalyzed by lumazine synthase from Aquifex aeolicus.
J.Mol.Biol., 328, 2003
|
|
2B99
 
 | | Crystal Structure of an archaeal pentameric riboflavin synthase Complex with a Substrate analog inhibitor | | Descriptor: | 6,7-DIOXO-5H-8-RIBITYLAMINOLUMAZINE, Riboflavin synthase | | Authors: | Ramsperger, A, Augustin, M, Schott, A.K, Gerhardt, S, Krojer, T, Eisenreich, W, Illarionov, B, Cushman, M, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2005-10-11 | | Release date: | 2005-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal Structure of an Archaeal Pentameric Riboflavin Synthase in Complex with a Substrate Analog Inhibitor: stereochemical implications
J.Biol.Chem., 281, 2006
|
|
2I0F
 
 | | Lumazine synthase RibH1 from Brucella abortus (Gene BruAb1_0785, Swiss-Prot entry Q57DY1) | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase 1, CALCIUM ION | | Authors: | Klinke, S, Zylberman, V, Bonomi, H.R, Haase, I, Guimaraes, B.G, Braden, B.C, Bacher, A, Fischer, M, Goldbaum, F.A. | | Deposit date: | 2006-08-10 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural and Kinetic Properties of Lumazine Synthase Isoenzymes in the Order Rhizobiales
J.Mol.Biol., 373, 2007
|
|
4KQ6
 
 | | Product complex of lumazine synthase from candida glabrata | | Descriptor: | 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, 6,7-dimethyl-8-ribityllumazine synthase, GLYCEROL, ... | | Authors: | Shankar, M, Wilbanks, S.M, Nakatani, Y, Monk, B.C, Tyndall, J.D.A. | | Deposit date: | 2013-05-14 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Catalysis product captured in lumazine synthase from the fungal pathogen Candida glabrata.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2F59
 
 | | Lumazine synthase RibH1 from Brucella abortus (Gene BruAb1_0785, Swiss-Prot entry Q57DY1) complexed with inhibitor 5-Nitro-6-(D-Ribitylamino)-2,4(1H,3H) Pyrimidinedione | | Descriptor: | 5-NITRO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, 6,7-dimethyl-8-ribityllumazine synthase 1, CALCIUM ION | | Authors: | Klinke, S, Zylberman, V, Bonomi, H.R, Haase, I, Guimaraes, B.G, Braden, B.C, Bacher, A, Fischer, M, Goldbaum, F.A. | | Deposit date: | 2005-11-25 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Kinetic Properties of Lumazine Synthase Isoenzymes in the Order Rhizobiales
J.Mol.Biol., 373, 2007
|
|
2VI5
 
 | | LUMAZINE SYNTHASE FROM MYCOBACTERIUM TUBERCULOSIS BOUND TO N-6-(ribitylamino)pyrimidine-2,4(1H,3H)-dione-5-yl-propionamide | | Descriptor: | 1-deoxy-1-{[(5S)-2,6-dioxo-5-(propanoylamino)-1,2,5,6-tetrahydropyrimidin-4-yl]amino}-D-ribitol, 6,7-DIMETHYL-8-RIBITYLLUMAZINE SYNTHASE, PHOSPHATE ION, ... | | Authors: | Morgunova, E, Zhang, Y, Jin, G, Illarionov, B, Bacher, A, Fischer, M, Cushman, M, Ladenstein, R. | | Deposit date: | 2007-11-27 | | Release date: | 2008-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A New Series of N-[2,4-Dioxo-6-D-Ribitylamino-1,2, 3,4-Tetrahydropyrimidin-5-Yl]Oxalamic Acid Derivatives as Inhibitors of Lumazine Syntase and Riboflavin Synthase: Design, Synthesis, Biochemical Evaluation, Crystallography and Mechanistic Implications.
J.Org.Chem., 73, 2008
|
|
2B98
 
 | | Crystal Structure of an archaeal pentameric riboflavin synthase | | Descriptor: | Riboflavin synthase | | Authors: | Ramsperger, A, Augustin, M, Schott, A.K, Gerhardt, S, Krojer, T, Eisenreich, W, Illarionov, B, Cushman, M, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2005-10-11 | | Release date: | 2005-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of an Archaeal Pentameric Riboflavin Synthase in Complex with a Substrate Analog Inhibitor: stereochemical implications
J.Biol.Chem., 281, 2006
|
|
1W29
 
 | | Lumazine Synthase from Mycobacterium tuberculosis bound to 3-(1,3,7- trihydro-9-D-ribityl-2,6,8-purinetrione-7-yl)butane 1-phosphate | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, 4-{2,6,8-TRIOXO-9-[(2R,3S,4R)-2,3,4,5-TETRAHYDROXYPENTYL]-1,2,3,6,8,9-HEXAHYDRO-7H-PURIN-7-YL}BUTYL DIHYDROGEN PHOSPHATE, 4-{2,6,8-TRIOXO-9-[(2S,3R,4R)-2,3,4,5-TETRAHYDROXYPENTYL]-1,2,3,6,8,9-HEXAHYDRO-7H-PURIN-7-YL}BUTYL DIHYDROGEN PHOSPHATE, ... | | Authors: | Morgunova, E, Meining, W, Illarionov, B, Haase, I, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2004-07-01 | | Release date: | 2005-03-03 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Lumazine Synthase from Mycobacterium Tuberculosis as a Target for Rational Drug Design: Binding Mode of a New Class of Purinetrione Inhibitors(,)
Biochemistry, 44, 2005
|
|
7X7M
 
 | | Lumazine Synthase from Aquifex aeolicus | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase | | Authors: | Sobhy, M.A, Hamdan, S.M. | | Deposit date: | 2022-03-09 | | Release date: | 2022-10-12 | | Last modified: | 2022-10-19 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Cryo-electron structures of the extreme thermostable enzymes Sulfur Oxygenase Reductase and Lumazine Synthase.
Plos One, 17, 2022
|
|
1KYY
 
 | | Lumazine Synthase from S.pombe bound to nitropyrimidinedione | | Descriptor: | 5-NITRO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, 6,7-Dimethyl-8-ribityllumazine Synthase, PHOSPHATE ION | | Authors: | Gerhardt, S, Haase, I, Steinbacher, S, Kaiser, J.T, Cushman, M, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2002-02-06 | | Release date: | 2002-07-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural basis of riboflavin binding to Schizosaccharomyces pombe 6,7-dimethyl-8-ribityllumazine synthase.
J.Mol.Biol., 318, 2002
|
|
1KYV
 
 | | Lumazine Synthase from S.pombe bound to riboflavin | | Descriptor: | 6,7-Dimethyl-8-ribityllumazine Synthase, PHOSPHATE ION, RIBOFLAVIN | | Authors: | Gerhardt, S, Haase, I, Steinbacher, S, Kaiser, J.T, Cushman, M, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2002-02-06 | | Release date: | 2002-07-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural basis of riboflavin binding to Schizosaccharomyces pombe 6,7-dimethyl-8-ribityllumazine synthase.
J.Mol.Biol., 318, 2002
|
|
1RVV
 
 | | SYNTHASE/RIBOFLAVIN SYNTHASE COMPLEX OF BACILLUS SUBTILIS | | Descriptor: | 5-NITRO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, PHOSPHATE ION, RIBOFLAVIN SYNTHASE | | Authors: | Ritsert, K, Huber, R, Turk, D, Ladenstein, R, Schmidt-Baese, K, Bacher, A. | | Deposit date: | 1995-10-25 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Studies on the lumazine synthase/riboflavin synthase complex of Bacillus subtilis: crystal structure analysis of reconstituted, icosahedral beta-subunit capsids with bound substrate analogue inhibitor at 2.4 A resolution.
J.Mol.Biol., 253, 1995
|
|
2OBX
 
 | | Lumazine synthase RibH2 from Mesorhizobium loti (Gene mll7281, Swiss-Prot entry Q986N2) complexed with inhibitor 5-Nitro-6-(D-Ribitylamino)-2,4(1H,3H) Pyrimidinedione | | Descriptor: | 5-NITRO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, 6,7-dimethyl-8-ribityllumazine synthase 1, PHOSPHATE ION | | Authors: | Klinke, S, Zylberman, V, Bonomi, H.R, Haase, I, Guimaraes, B.G, Braden, B.C, Bacher, A, Fischer, M, Goldbaum, F.A. | | Deposit date: | 2006-12-20 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural and kinetic properties of lumazine synthase isoenzymes in the order rhizobiales
J.Mol.Biol., 373, 2007
|
|
8F25
 
 | |
