7Y9N
 
 | | an engineered 5-helix bundle derived from SARS-CoV-2 S2 in complex with HR2P | | Descriptor: | SARS-coV-2 S2 subunit, Spike protein S2',5HB-H2 | | Authors: | Lu, G.W, Lin, X, Guo, L.Y, Lin, S. | | Deposit date: | 2022-06-25 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.885 Å) | | Cite: | An engineered 5-helix bundle derived from SARS-CoV-2 S2 pre-binds sarbecoviral spike at both serological- and endosomal-pH to inhibit virus entry.
Emerg Microbes Infect, 11, 2022
|
|
7ZBU
 
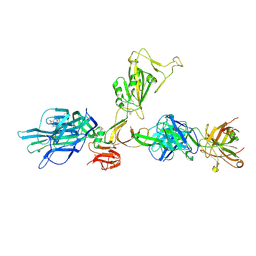 | | CryoEM structure of SARS-CoV-2 spike monomer in complex with neutralising antibody P008_60 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-2-[[5-[(3-ethyl-4-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, P008_60 antibody, ... | | Authors: | Rosa, A, Pye, V.E, Cronin, N, Cherepanov, P. | | Deposit date: | 2022-03-24 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.31 Å) | | Cite: | A neutralizing epitope on the SD1 domain of SARS-CoV-2 spike targeted following infection and vaccination.
Cell Rep, 40, 2022
|
|
7RU1
 
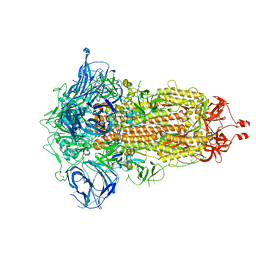 | | SARS-CoV-2-6P-Mut7 S protein (C3 symmetry) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Ozorowski, G, Turner, H.L, Ward, A.B. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Engineering SARS-CoV-2 neutralizing antibodies for increased potency and reduced viral escape pathways.
Iscience, 25, 2022
|
|
7RU4
 
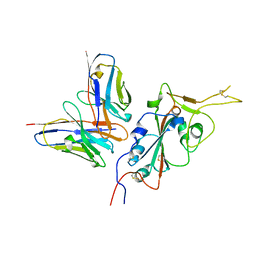 | | CC6.33 IgG in complex with SARS-CoV-2-6P-Mut7 S protein (RBD/Fv local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CC6.33 IgG heavy chain Fv, ... | | Authors: | Ozorowski, G, Turner, H.L, Ward, A.B. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Engineering SARS-CoV-2 neutralizing antibodies for increased potency and reduced viral escape pathways.
Iscience, 25, 2022
|
|
7RU5
 
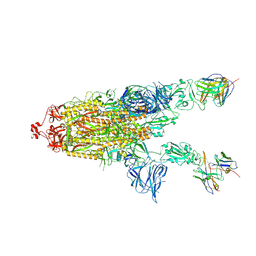 | | CC6.30 fragment antigen binding in complex with SARS-CoV-2-6P-Mut7 S protein (non-uniform refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CC6.30 Fab heavy chain Fv, ... | | Authors: | Ozorowski, G, Turner, H.L, Ward, A.B. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Engineering SARS-CoV-2 neutralizing antibodies for increased potency and reduced viral escape pathways.
Iscience, 25, 2022
|
|
7RU8
 
 | | CC6.30 fragment antigen binding in complex with SARS-CoV-2-6P-Mut7 S protein (RBD/Fv local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CC6.30 Fab Kappa chain Fv, ... | | Authors: | Ozorowski, G, Turner, H.L, Ward, A.B. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Engineering SARS-CoV-2 neutralizing antibodies for increased potency and reduced viral escape pathways.
Iscience, 25, 2022
|
|
7T3M
 
 | | SARS-CoV-2 S (Spike Glycoprotein) D614G with Three (3) RBDs Up, Bound to Antibody 2-7 scFv, composite map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 2-7 scFv, ... | | Authors: | Byrne, P.O, McLellan, J.S. | | Deposit date: | 2021-12-08 | | Release date: | 2022-08-24 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | IgG-like bispecific antibodies with potent and synergistic neutralization against circulating SARS-CoV-2 variants of concern.
Nat Commun, 13, 2022
|
|
7T67
 
 | | SARS-CoV-2 S (Spike Glycoprotein) D614G with One(1) RBD Up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Byrne, P.O, McLellan, J.S. | | Deposit date: | 2021-12-13 | | Release date: | 2022-08-24 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | IgG-like bispecific antibodies with potent and synergistic neutralization against circulating SARS-CoV-2 variants of concern.
Nat Commun, 13, 2022
|
|
7YDI
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs and 3 ACE2, focused refinement of RBD region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32, Light chain of R1-32, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-04 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YE9
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-05 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YEG
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs and 3 ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-05 | | Release date: | 2022-08-24 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
8DAD
 
 | | SARS-CoV-2 receptor binding domain in complex with AZ090 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AZ090 Fab Heavy Chain, AZ090 Fab Light Chain, ... | | Authors: | Zong, S, Wang, Z, Gaebler, C, Nussenzweig, M. | | Deposit date: | 2022-06-13 | | Release date: | 2022-08-24 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | SARS-CoV-2 receptor binding domain in complex with AZ090 Fab
To Be Published
|
|
8DI5
 
 | | Cryo-EM structure of SARS-CoV-2 Beta (B.1.351) spike protein in complex with VH domain F6 (focused refinement of RBD and VH F6) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, VH F6 | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Subramaniam, S. | | Deposit date: | 2022-06-28 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Potent and broad neutralization of SARS-CoV-2 variants of concern (VOCs) including omicron sub-lineages BA.1 and BA.2 by biparatopic human VH domains.
Iscience, 25, 2022
|
|
8DOY
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with an inhibitor TKB-198 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3C-like proteinase nsp5, ... | | Authors: | Bulut, H, Hayashi, H, Tsuji, K, Kuwata, N, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2022-07-14 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Potent and biostable inhibitors of the main protease of SARS-CoV-2.
Iscience, 25, 2022
|
|
8DZ9
 
 | | Crystal Structure of SARS-CoV-2 Main protease G143S mutant in complex with Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Noske, G.D, Oliva, G, Godoy, A.S. | | Deposit date: | 2022-08-06 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.664 Å) | | Cite: | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
8E1Y
 
 | | Crystal Structure of SARS-CoV-2 Main protease A193S mutant in complex with Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Noske, G.D, Oliva, G, Godoy, A.S. | | Deposit date: | 2022-08-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
7VBD
 
 | | Crystal structure of SARS-Cov-2 nucleocapsid N-terminal domain (NTD) protein,pH8.0 | | Descriptor: | Nucleoprotein | | Authors: | Zeng, P, Zhou, X.L, Zhong, F.L, Li, J, Zhang, J. | | Deposit date: | 2021-08-31 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of SARS-Cov-2 nucleocapsid N-terminal domain (NTD) protein,pH8.0
To Be Published
|
|
7WBZ
 
 | | Crystal structure of the SARS-Cov-2 RBD in complex with Fab 2303 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2303 heavy chain, 2303 light chain, ... | | Authors: | Xiang, Y, Ma, B. | | Deposit date: | 2021-12-17 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Conformational flexibility in neutralization of SARS-CoV-2 by naturally elicited anti-SARS-CoV-2 antibodies.
Commun Biol, 5, 2022
|
|
7XCH
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein (S-6P-RRAR) in complex with human ACE2 ectodomain (two-RBD-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Gao, G.F, Qi, J.X, Zhao, Z.N, Liu, S, Xie, Y.F. | | Deposit date: | 2022-03-24 | | Release date: | 2022-08-31 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
7XCI
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron RBD in complex with human ACE2 ectodomain (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Gao, G.F, Qi, J.X, Zhao, Z.N, Liu, S, Xie, Y.F. | | Deposit date: | 2022-03-24 | | Release date: | 2022-08-31 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
7XCK
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron RBD in complex with S309 fab (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, S309 heavy chain, S309 light chain, ... | | Authors: | Gao, G.F, Qi, J.X, Zhao, Z.N, Xie, Y.F, Liu, S. | | Deposit date: | 2022-03-24 | | Release date: | 2022-08-31 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
7XCP
 
 | | Cryo-EM structure of Omicron RBD complexed with ACE2 and 304 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 304 Fab, ... | | Authors: | Zhao, Z, Qi, J, Gao, F.G. | | Deposit date: | 2022-03-24 | | Release date: | 2022-08-31 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
7YA1
 
 | | Cryo-EM structure of hACE2-bound SARS-CoV-2 Omicron spike protein with L371S, P373S and F375S mutations (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-06-27 | | Release date: | 2022-08-31 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7YAD
 
 | | Cryo-EM structure of S309-RBD-RBD-S309 in the S309-bound Omicron spike protein (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, S309 neutralizing antibody heavy chain, S309 neutralizing antibody light chain, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, F. | | Deposit date: | 2022-06-27 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
8DDI
 
 | |








