7JX6
 
 | |
7JZL
 
 | |
7JZM
 
 | |
7JZN
 
 | |
7JZU
 
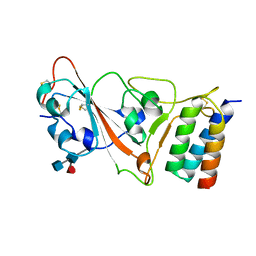 | |
5RL9
 
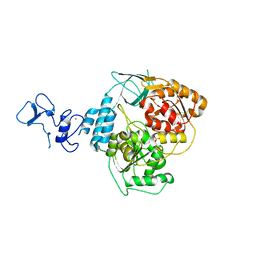 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z1703168683 | | Descriptor: | 1-(3-fluoro-4-methylphenyl)methanesulfonamide, Helicase, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.788 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
7CWC
 
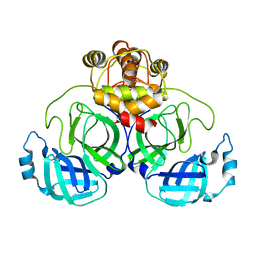 | |
7K3N
 
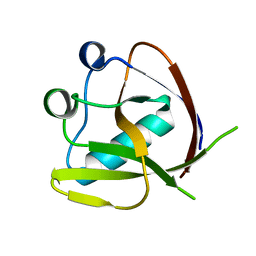 | | Crystal Structure of NSP1 from SARS-CoV-2 | | Descriptor: | Host translation inhibitor nsp1 | | Authors: | Semper, C, Watanabe, N, Chang, C, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-11 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural characterization of nonstructural protein 1 from SARS-CoV-2.
Iscience, 24, 2021
|
|
7K7P
 
 | |
7D47
 
 | | Crystal structure of SARS-CoV-2 Papain-like protease C111S | | Descriptor: | CALCIUM ION, Non-structural protein 3, ZINC ION | | Authors: | Wu, K.-P, Chen, S.-K, Lu, Y.-C, Huang, Y.-C.J, Lee, M.-H. | | Deposit date: | 2020-09-22 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of SARS-CoV-2 Papain-like protease
To Be Published
|
|
7JOY
 
 | |
7JWB
 
 | |
7K43
 
 | |
7K45
 
 | |
7K4N
 
 | |
6XKP
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody CV07-270 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CV07-270 Heavy Chain, CV07-270 Light Chain, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-06-26 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | A Therapeutic Non-self-reactive SARS-CoV-2 Antibody Protects from Lung Pathology in a COVID-19 Hamster Model.
Cell, 183, 2020
|
|
6XKQ
 
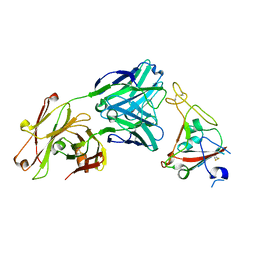 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody CV07-250 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CV07-250 Heavy Chain, CV07-250 Light Chain, ... | | Authors: | Yuan, M, Liu, H, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-06-26 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A Therapeutic Non-self-reactive SARS-CoV-2 Antibody Protects from Lung Pathology in a COVID-19 Hamster Model.
Cell, 183, 2020
|
|
6ZPE
 
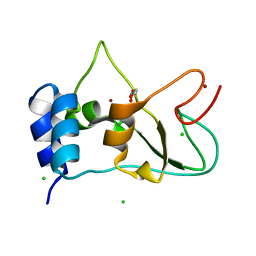 | | Nonstructural protein 10 (nsp10) from SARS CoV-2 | | Descriptor: | CHLORIDE ION, GLYCEROL, Replicase polyprotein 1ab, ... | | Authors: | Fisher, S.Z, Kozielski, F. | | Deposit date: | 2020-07-08 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of Non-Structural Protein 10 from Severe Acute Respiratory Syndrome Coronavirus-2.
Int J Mol Sci, 21, 2020
|
|
7JMW
 
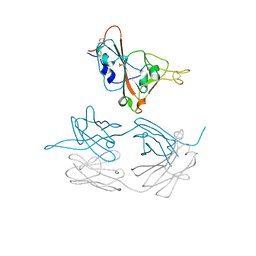 | | Crystal structure of SARS-CoV-2 spike protein receptor-binding domain in complex with cross-neutralizing antibody COVA1-16 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVA1-16 heavy chain, COVA1-16 light chain, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-08-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Cross-Neutralization of a SARS-CoV-2 Antibody to a Functionally Conserved Site Is Mediated by Avidity.
Immunity, 53, 2020
|
|
7JV2
 
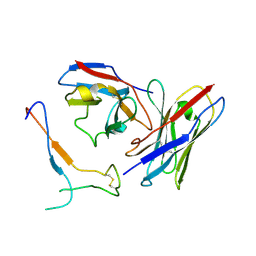 | | SARS-CoV-2 spike in complex with the S2H13 neutralizing antibody Fab fragment (local refinement of the receptor-binding motif and Fab variable domains) | | Descriptor: | S2H13 Fab heavy chain, S2H13 Fab light chain, Spike glycoprotein | | Authors: | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2020-08-20 | | Release date: | 2020-10-14 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7JV6
 
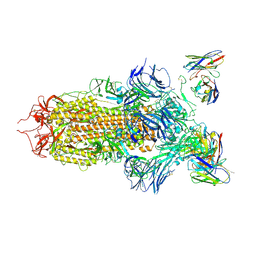 | | SARS-CoV-2 spike in complex with the S2H13 neutralizing antibody (closed conformation) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S2H13 Fab heavy chain, ... | | Authors: | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2020-08-20 | | Release date: | 2020-10-14 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7JVA
 
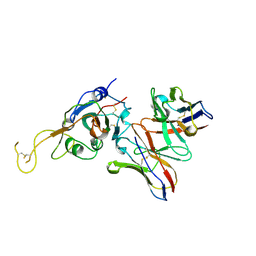 | | SARS-CoV-2 spike in complex with the S2A4 neutralizing antibody Fab fragment (local refinement of the receptor-binding domain and Fab variable domains) | | Descriptor: | S2A4 Fab heavy chain, S2A4 Fab light chain, Spike glycoprotein, ... | | Authors: | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2020-08-20 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7JVC
 
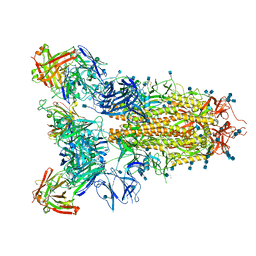 | | SARS-CoV-2 spike in complex with the S2A4 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S2A4 Fab heavy chain, ... | | Authors: | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2020-08-20 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7JW0
 
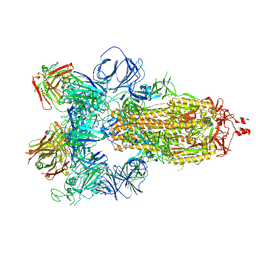 | | SARS-CoV-2 spike in complex with the S304 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S304 Fab heavy chain, ... | | Authors: | Walls, A.C, Park, Y.J, Tortorici, M.A, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2020-08-24 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7JX3
 
 | | Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab domain of monoclonal antibody S2H14, Heavy chain of Fab domain of monoclonal antibody S304, ... | | Authors: | Snell, G, Czudnochowski, N, Rosen, L.E, Nix, J.C, Corti, D, Veesler, D, Park, Y.J, Walls, A.C, Tortorici, M.A, Cameroni, E, Pinto, D, Beltramello, M, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-08-26 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|








