-検索条件
-検索結果
検索 (著者・登録者: van & heel & m)の結果全46件を表示しています

EMDB-28013: 
Cryo-EM structure of the Glutaminase C core filament (fGAC)

PDB-8ec6: 
Cryo-EM structure of the Glutaminase C core filament (fGAC)

EMDB-26481: 
Cryo-EM Structure of Bl_Man38A nucleophile mutant in complex with mannose at 2.7 A

PDB-7ufu: 
Cryo-EM Structure of Bl_Man38A nucleophile mutant in complex with mannose at 2.7 A

EMDB-26480: 
Cryo-EM Structure of Bl_Man38C at 2.9 A

PDB-7uft: 
Cryo-EM Structure of Bl_Man38C at 2.9 A

EMDB-26479: 
Cryo-EM Structure of Bl_Man38B at 3.4 A

PDB-7ufs: 
Cryo-EM Structure of Bl_Man38B at 3.4 A

EMDB-26478: 
Cryo-EM Structure of Bl_Man38A at 2.7 A

PDB-7ufr: 
Cryo-EM Structure of Bl_Man38A at 2.7 A

EMDB-23698: 
Human Septin Hexameric Complex SEPT2G/SEPT6/SEPT7 by Single Particle Cryo-EM

PDB-7m6j: 
Human Septin Hexameric Complex SEPT2G/SEPT6/SEPT7 by Single Particle Cryo-EM

PDB-7ko8: 
Cryo-EM structure of the mature and infective Mayaro virus

EMDB-22961: 
Cryo-EM structure of the mature and infective Mayaro virus

PDB-5m3l: 
Single-particle cryo-EM using alignment by classification (ABC): the structure of Lumbricus terrestris hemoglobin

EMDB-3434: 
Single-particle cryo-EM using alignment by classification (ABC):Lumbricus terrestris hemoglobin at near-atomic resolution

EMDB-2825: 
Cryo-EM structure of worm hemoglobin

EMDB-2459: 
Structure and Host Adhesion Mechanism of Virulent Lactococcal Phage p2

EMDB-2462: 
Structure and Host Adhesion Mechanism of Virulent Lactococcal Phage p2

EMDB-2463: 
The electron cryo-miscroscopy reconstruction of the connector of lactococcal phage p2
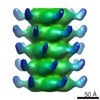
EMDB-2464: 
The electron miscroscopy reconstruction of the tail of lactococcal phage p2

EMDB-2133: 
The Structure of Lactococcal Phage TP901-1 by electron microscopy: the capsid

EMDB-2227: 
The Structure of Lactococcal Phage TP901-1 by electron microscopy: the connector

EMDB-2228: 
The Structure of Lactococcal Phage TP901-1 by electron microscopy: the helical tail
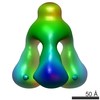
EMDB-1792: 
EM map of TP901-1 BppU-BppL complex

EMDB-1793: 
The three-dimensional reconstruction of the baseplate of wild-type TP901-1
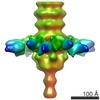
EMDB-1794: 
The three-dimensional reconstruction of the baseplate of TP901-1 bppL mutant

EMDB-1795: 
The three-dimensional reconstruction of the baseplate of TP901-1 bppU mutant
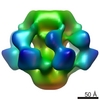
EMDB-1706: 
Cryo-EM map of Lactococcal phage p2 baseplate consisting of ORF 15, 16 and 18.

EMDB-1699: 
Structure of Lactococcal Phage p2 Baseplate and its Mechanism of Activation

EMDB-1552: 
Molecular Architecture of the 'stressosome', a signal transduction hub

EMDB-1555: 
Molecular Architecture of the 'stressosome', a signal transduction hub

EMDB-1556: 
Molecular Architecture of the 'stressosome', a signal transduction hub

EMDB-1558: 
Molecular Architecture of the 'stressosome', a signal transduction hub

EMDB-1064: 
Visualization of release factor 3 on the ribosome during termination of protein synthesis.

EMDB-1065: 
Visualization of release factor 3 on the ribosome during termination of protein synthesis.

EMDB-1005: 
Structure of the Escherichia coli ribosomal termination complex with release factor 2.

EMDB-1004: 
Ribosome interactions of aminoacyl-tRNA and elongation factor Tu in the codon-recognition complex.

EMDB-1020: 
Structure of a viral DNA gatekeeper at 10 A resolution by cryo-electron microscopy.

EMDB-1021: 
Structure of a viral DNA gatekeeper at 10 A resolution by cryo-electron microscopy.

PDB-1ml5: 
Structure of the E. coli ribosomal termination complex with release factor 2

EMDB-1019: 
The Escherichia coli large ribosomal subunit at 7.5 A resolution.

PDB-1mj1: 
FITTING THE TERNARY COMPLEX OF EF-Tu/tRNA/GTP AND RIBOSOMAL PROTEINS INTO A 13 A CRYO-EM MAP OF THE COLI 70S RIBOSOME

PDB-1c2w: 
23S RRNA STRUCTURE FITTED TO A CRYO-ELECTRON MICROSCOPIC MAP AT 7.5 ANGSTROMS RESOLUTION
PDB-1c2x: 
5S RRNA STRUCTURE FITTED TO A CRYO-ELECTRON MICROSCOPIC MAP AT 7.5 ANGSTROMS RESOLUTION

PDB-487d: 
SEVEN RIBOSOMAL PROTEINS FITTED TO A CRYO-ELECTRON MICROSCOPIC MAP OF THE LARGE 50S SUBUNIT AT 7.5 ANGSTROMS RESOLUTION
 ムービー
ムービー コントローラー
コントローラー 構造ビューア
構造ビューア EMN検索について
EMN検索について



 wwPDBはEMDBデータモデルのバージョン3へ移行します
wwPDBはEMDBデータモデルのバージョン3へ移行します
