[English] 日本語
 Yorodumi
Yorodumi- EMDB-1019: The Escherichia coli large ribosomal subunit at 7.5 A resolution. -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1019 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
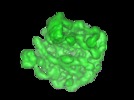
| Title | The Escherichia coli large ribosomal subunit at 7.5 A resolution. | |||||||||
 Map data Map data | The Escherichia coli large ribosomal subunit at 7.5 A resolution. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 7.5 Å | |||||||||
 Authors Authors | Matadeen R | |||||||||
 Citation Citation |  Journal: Structure / Year: 1999 Journal: Structure / Year: 1999Title: The Escherichia coli large ribosomal subunit at 7.5 A resolution. Authors: R Matadeen / A Patwardhan / B Gowen / E V Orlova / T Pape / M Cuff / F Mueller / R Brimacombe / M van Heel /  Abstract: BACKGROUND: In recent years, the three-dimensional structure of the ribosome has been visualised in different functional states by single-particle cryo-electron microscopy (cryo-EM) at 13-25 A ...BACKGROUND: In recent years, the three-dimensional structure of the ribosome has been visualised in different functional states by single-particle cryo-electron microscopy (cryo-EM) at 13-25 A resolution. Even more recently, X-ray crystallography has achieved resolution levels better than 10 A for the ribosomal structures of thermophilic and halophilic organisms. We present here the 7.5 A solution structure of the 50S large subunit of the Escherichia coli ribosome, as determined by cryo-EM and angular reconstitution. RESULTS: The reconstruction reveals a host of new details including the long alpha helix connecting the N- and C-terminal domains of the L9 protein, which is found wrapped like a collar around the ...RESULTS: The reconstruction reveals a host of new details including the long alpha helix connecting the N- and C-terminal domains of the L9 protein, which is found wrapped like a collar around the base of the L1 stalk. A second L7/L12 dimer is now visible below the classical L7/L12 'stalk', thus revealing the position of the entire L8 complex. Extensive conformational changes occur in the 50S subunit upon 30S binding; for example, the L9 protein moves by some 50 A. Various rRNA stem-loops are found to be involved in subunit binding: helix h38, located in the A-site finger; h69, on the rim of the peptidyl transferase centre cleft; and h34, in the principal interface protrusion. CONCLUSIONS: Single-particle cryo-EM is rapidly evolving towards the resolution levels required for the direct atomic interpretation of the structure of the ribosome. Structural details such as the ...CONCLUSIONS: Single-particle cryo-EM is rapidly evolving towards the resolution levels required for the direct atomic interpretation of the structure of the ribosome. Structural details such as the minor and major grooves in rRNA double helices and alpha helices of the ribosomal proteins can already be visualised directly in cryo-EM reconstructions of ribosomes frozen in different functional states. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1019.map.gz emd_1019.map.gz | 12.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1019-v30.xml emd-1019-v30.xml emd-1019.xml emd-1019.xml | 8.7 KB 8.7 KB | Display Display |  EMDB header EMDB header |
| Images |  1019.gif 1019.gif | 20.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1019 http://ftp.pdbj.org/pub/emdb/structures/EMD-1019 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1019 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1019 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_1019.map.gz / Format: CCP4 / Size: 20.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1019.map.gz / Format: CCP4 / Size: 20.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The Escherichia coli large ribosomal subunit at 7.5 A resolution. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : 50S E.coli ribosomal subunit
| Entire | Name: 50S E.coli ribosomal subunit |
|---|---|
| Components |
|
-Supramolecule #1000: 50S E.coli ribosomal subunit
| Supramolecule | Name: 50S E.coli ribosomal subunit / type: sample / ID: 1000 / Oligomeric state: single entity / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 1.5 MDa / Theoretical: 1.5 MDa / Method: Sedimentation |
-Supramolecule #1: 50S ribosomal subunit
| Supramolecule | Name: 50S ribosomal subunit / type: complex / ID: 1 / Recombinant expression: No / Ribosome-details: ribosome-prokaryote: LSU 50S |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 1.5 MDa / Theoretical: 1.5 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 20mM HEPES-KOH 6 mM MgCl2, 150 mM NH4Cl |
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER / Details: Vitrification instrument: in-house freeze plunger / Method: Blot for 2 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM200FEG/UT |
|---|---|
| Temperature | Average: 96 K |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: PATCHWORK DENSITOMETER / Digitization - Sampling interval: 5 µm / Number real images: 7 / Average electron dose: 10 e/Å2 / Bits/pixel: 16 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 37600 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 38000 |
| Sample stage | Specimen holder: eucentric / Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| CTF correction | Details: Each particle |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 7.5 Å / Resolution method: FSC 3 SIGMA CUT-OFF / Software - Name: Imagic / Number images used: 16000 |
| Final two d classification | Number classes: 1000 |
-Atomic model buiding 1
| Software | Name: ESSENS |
|---|
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















