+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the Glutaminase C core filament (fGAC) | |||||||||
 Map data Map data | Local refinement, after helical refinement with D2 symmetry expansion | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Mitochondria / Filament / HYDROLASE | |||||||||
| Function / homology | glutaminase / Isoform 2 of Glutaminase kidney isoform, mitochondrial Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
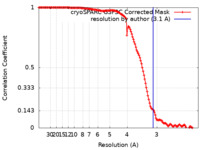
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Ambrosio AL / Dias SM / Quesnay JE / Portugal RV / Cassago A / van Heel MG / Islam Z / Rodrigues CT | |||||||||
| Funding support |  Brazil, 1 items Brazil, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2023 Journal: Nat Struct Mol Biol / Year: 2023Title: Molecular mechanism of glutaminase activation through filamentation and the role of filaments in mitophagy protection. Authors: Douglas Adamoski / Marilia Meira Dias / Jose Edwin Neciosup Quesñay / Zhengyi Yang / Ievgeniia Zagoriy / Anna M Steyer / Camila Tanimoto Rodrigues / Alliny Cristiny da Silva Bastos / Bianca ...Authors: Douglas Adamoski / Marilia Meira Dias / Jose Edwin Neciosup Quesñay / Zhengyi Yang / Ievgeniia Zagoriy / Anna M Steyer / Camila Tanimoto Rodrigues / Alliny Cristiny da Silva Bastos / Bianca Novaes da Silva / Renna Karoline Eloi Costa / Flávia Mayumi Odahara de Abreu / Zeyaul Islam / Alexandre Cassago / Marin Gerard van Heel / Sílvio Roberto Consonni / Simone Mattei / Julia Mahamid / Rodrigo Villares Portugal / Andre Luis Berteli Ambrosio / Sandra Martha Gomes Dias /     Abstract: Glutaminase (GLS), which deaminates glutamine to form glutamate, is a mitochondrial tetrameric protein complex. Although inorganic phosphate (Pi) is known to promote GLS filamentation and activation, ...Glutaminase (GLS), which deaminates glutamine to form glutamate, is a mitochondrial tetrameric protein complex. Although inorganic phosphate (Pi) is known to promote GLS filamentation and activation, the molecular basis of this mechanism is unknown. Here we aimed to determine the molecular mechanism of Pi-induced mouse GLS filamentation and its impact on mitochondrial physiology. Single-particle cryogenic electron microscopy revealed an allosteric mechanism in which Pi binding at the tetramer interface and the activation loop is coupled to direct nucleophile activation at the active site. The active conformation is prone to enzyme filamentation. Notably, human GLS filaments form inside tubulated mitochondria following glutamine withdrawal, as shown by in situ cryo-electron tomography of cells thinned by cryo-focused ion beam milling. Mitochondria with GLS filaments exhibit increased protection from mitophagy. We reveal roles of filamentous GLS in mitochondrial morphology and recycling. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28013.map.gz emd_28013.map.gz | 230 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28013-v30.xml emd-28013-v30.xml emd-28013.xml emd-28013.xml | 20 KB 20 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28013_fsc.xml emd_28013_fsc.xml | 13.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_28013.png emd_28013.png | 50.8 KB | ||
| Filedesc metadata |  emd-28013.cif.gz emd-28013.cif.gz | 6.6 KB | ||
| Others |  emd_28013_half_map_1.map.gz emd_28013_half_map_1.map.gz emd_28013_half_map_2.map.gz emd_28013_half_map_2.map.gz | 226.9 MB 226.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28013 http://ftp.pdbj.org/pub/emdb/structures/EMD-28013 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28013 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28013 | HTTPS FTP |
-Validation report
| Summary document |  emd_28013_validation.pdf.gz emd_28013_validation.pdf.gz | 814.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_28013_full_validation.pdf.gz emd_28013_full_validation.pdf.gz | 813.9 KB | Display | |
| Data in XML |  emd_28013_validation.xml.gz emd_28013_validation.xml.gz | 21.6 KB | Display | |
| Data in CIF |  emd_28013_validation.cif.gz emd_28013_validation.cif.gz | 28 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28013 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28013 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28013 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28013 | HTTPS FTP |
-Related structure data
| Related structure data |  8ec6MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28013.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28013.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local refinement, after helical refinement with D2 symmetry expansion | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half-map A
| File | emd_28013_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map B
| File | emd_28013_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Glutaminase C filament, bound to inorganic phosphate
| Entire | Name: Glutaminase C filament, bound to inorganic phosphate |
|---|---|
| Components |
|
-Supramolecule #1: Glutaminase C filament, bound to inorganic phosphate
| Supramolecule | Name: Glutaminase C filament, bound to inorganic phosphate / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 0.53 kDa/nm |
-Macromolecule #1: Isoform 2 of Glutaminase kidney isoform, mitochondrial
| Macromolecule | Name: Isoform 2 of Glutaminase kidney isoform, mitochondrial type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO / EC number: glutaminase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 53.326961 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSHMVAAGDN KIKQGLLPSL EDLLFYTIAE GQEKIPVHKF ITALKSTGLR TSDPRLKECM DMLRLTLQTT SDGVMLDKDL FKKCVQSNI VLLTQAFRRK FVIPDFMSFT SHIDELYESA KKQSGGKVAD YIPQLAKFSP DLWGVSVCTV DGQRHSIGDT K VPFCLQSC ...String: GSHMVAAGDN KIKQGLLPSL EDLLFYTIAE GQEKIPVHKF ITALKSTGLR TSDPRLKECM DMLRLTLQTT SDGVMLDKDL FKKCVQSNI VLLTQAFRRK FVIPDFMSFT SHIDELYESA KKQSGGKVAD YIPQLAKFSP DLWGVSVCTV DGQRHSIGDT K VPFCLQSC VKPLKYAIAV NDLGTEYVHR YVGKEPSGLR FNKLFLNEDD KPHNPMVNAG AIVVTSLIKQ GVNNAEKFDY VM QFLNKMA GNEYVGFSNA TFQSERESGD RNFAIGYYLK EKKCFPEGTD MVGILDFYFQ LCSIEVTCES ASVMAATLAN GGF CPITGE RVLSPEAVRN TLSLMHSCGM YDFSGQFAFH VGLPAKSGVA GGILLVVPNV MGMMCWSPPL DKMGNSVKGI HFCH DLVSL CNFHNYDNLR HFAKKLDPRR EGGDQRHSFG PLDYESLQQE LALKDTVWKK VSPESSDDTS TTVVYRMESL GERS UniProtKB: Isoform 2 of Glutaminase kidney isoform, mitochondrial |
-Macromolecule #2: PHOSPHATE ION
| Macromolecule | Name: PHOSPHATE ION / type: ligand / ID: 2 / Number of copies: 8 / Formula: PO4 |
|---|---|
| Molecular weight | Theoretical: 94.971 Da |
| Chemical component information |  ChemComp-PO4: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 0.33 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8.5 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV | |||||||||||||||
| Details | In the presence of phosphate, GAC polymerizes and becomes polydisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 3.0 µm / Calibrated defocus min: 2.0 µm / Calibrated magnification: 59000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A / Chain - Residue range: 128-674 / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | run = *minimization_global *rigid_body local_grid_search *adp *occupancy *nqh_flips |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 162.1 / Target criteria: Correlation Coefficient |
| Output model |  PDB-8ec6: |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)