-Search query
-Search result
Showing all 46 items for (author: erramilli & s)

EMDB-43683: 
Cryo-EM structure of FLVCR2 in the inward-facing state with choline bound

EMDB-43684: 
Cryo-EM structure of FLVCR2 in the outward-facing state with choline bound

PDB-8vzn: 
Cryo-EM structure of FLVCR2 in the inward-facing state with choline bound

PDB-8vzo: 
Cryo-EM structure of FLVCR2 in the outward-facing state with choline bound
EMDB-40622: 
Chlorella virus Hyaluronan Synthase bound to GlcA extended GlcNAc primer

EMDB-40623: 
Chlorella virus Hyaluronan Synthase bound to GlcNAc primer and UDP-GlcA

EMDB-40624: 
Chlorella virus Hyaluronan Synthase bound to GlcA extended GlcNAc primer and UDP

EMDB-40591: 
Xenopus laevis hyaluronan synthase 1

EMDB-40594: 
Xenopus laevis hyaluronan synthase 1, nascent HA polymer bound state
EMDB-40598: 
Xenopus laevis hyaluronan synthase 1, UDP-bound, gating loop inserted state
EMDB-41255: 
Cryo-EM structure of Pseudomonas aeruginosa TonB-dependent transporter PhuR in complex with synthetic antibody and heme

PDB-8the: 
Cryo-EM structure of Pseudomonas aeruginosa TonB-dependent transporter PhuR in complex with synthetic antibody and heme

EMDB-41899: 
Cryo-EM structure of human claudin-4 complex with Clostridium perfringens enterotoxin C-terminal domain, sFab COP-1, and Nanobody (Loose Mask)

EMDB-41915: 
Cryo-EM structure of human claudin-4 complex with Clostridium perfringens enterotoxin C-terminal domain and sFab COP-1

EMDB-41897: 
Cryo-EM structure of human claudin-4 complex with Clostridium perfringens enterotoxin C-terminal domain, sFab COP-1, and Nanobody (Tight Mask)
EMDB-24698: 
Cryo-EM structure of the HIV-1 restriction factor human SERINC3

EMDB-24705: 
Human SERINC3-DeltaICL4

PDB-7ru6: 
Cryo-EM structure of the HIV-1 restriction factor human SERINC3

PDB-7rug: 
Human SERINC3-DeltaICL4

EMDB-25836: 
CryoEM Structure of sFab COP-3 Complex with human claudin-4 and Clostridium perfringens enterotoxin C-terminal domain focused map

EMDB-25750: 
Structure of the peroxisomal retro-translocon formed by a heterotrimeric ubiquitin ligase complex

PDB-7t92: 
Structure of the peroxisomal retro-translocon formed by a heterotrimeric ubiquitin ligase complex
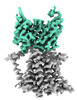
EMDB-26054: 
Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its ligand bound state
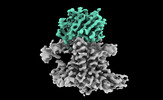
EMDB-26057: 
Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its apo state

PDB-7tpg: 
Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its ligand bound state

PDB-7tpj: 
Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its apo state

EMDB-25834: 
CryoEM Structure of sFab COP-2 Complex with human claudin-4 and Clostridium perfringens enterotoxin C-terminal domain

EMDB-25835: 
CryoEM Structure of sFab COP-3 Complex with human claudin-4 and Clostridium perfringens enterotoxin C-terminal domain

PDB-7tdm: 
CryoEM Structure of sFab COP-2 Complex with human claudin-4 and Clostridium perfringens enterotoxin C-terminal domain

PDB-7tdn: 
CryoEM Structure of sFab COP-3 Complex with human claudin-4 and Clostridium perfringens enterotoxin C-terminal domain

EMDB-12365: 
Nanodisc reconstituted human ABCB4 in complex with 4B1-Fab and QA2-Fab (apo-inward-open conformation)

EMDB-12366: 
Nanodisc reconstituted human ABCB4 in complex with 4B1-Fab and QA2-Fab (phosphatidylcholine-bound, occluded conformation)

EMDB-12367: 
Nanodisc reconstituted human ABCB4 in complex with 4B1-Fab (posaconazole-bound, inward-open conformation)

PDB-7niu: 
Nanodisc reconstituted human ABCB4 in complex with 4B1-Fab and QA2-Fab (apo-inward-open conformation)

PDB-7niv: 
Nanodisc reconstituted human ABCB4 in complex with 4B1-Fab and QA2-Fab (phosphatidylcholine-bound, occluded conformation)

PDB-7niw: 
Nanodisc reconstituted human ABCB4 in complex with 4B1-Fab (posaconazole-bound, inward-open conformation)

EMDB-23883: 
Single-Particle Cryo-EM Structure of Major Facilitator Superfamily Domain containing 2A in complex with LPC-18:3

PDB-7mjs: 
Single-Particle Cryo-EM Structure of Major Facilitator Superfamily Domain containing 2A in complex with LPC-18:3

EMDB-20857: 
CryoEM structure of human alpha4beta2 nicotinic acetylcholine receptor in complex with varenicline

EMDB-20863: 
CryoEM structure of human alpha4beta2 nicotinic acetylcholine receptor with varenicline in complex with anti-BRIL synthetic antibody BAK5

PDB-6ur8: 
CryoEM structure of human alpha4beta2 nicotinic acetylcholine receptor in complex with varenicline

PDB-6usf: 
CryoEM structure of human alpha4beta2 nicotinic acetylcholine receptor with varenicline in complex with anti-BRIL synthetic antibody BAK5

EMDB-20806: 
Single-Particle Cryo-EM Structure of Plasmodium falciparum Chloroquine Resistance Transporter (PfCRT) 7G8 Isoform

PDB-6ukj: 
Single-Particle Cryo-EM Structure of Plasmodium falciparum Chloroquine Resistance Transporter (PfCRT) 7G8 Isoform

EMDB-7517: 
Rhodopsin-Gi

PDB-6cmo: 
Rhodopsin-Gi complex
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
