[English] 日本語
 Yorodumi
Yorodumi- EMDB-25750: Structure of the peroxisomal retro-translocon formed by a heterot... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the peroxisomal retro-translocon formed by a heterotrimeric ubiquitin ligase complex | |||||||||
 Map data Map data | Structure of the peroxisomal retro-translocon formed by a heterotrimeric ubiquitin ligase complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | peroxisome / retro-translocon / ubiquitin ligase / TRANSLOCASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationperoxisomal importomer complex / RING-type E3 ubiquitin transferase (cysteine targeting) / protein import into peroxisome matrix, receptor recycling / peroxisomal membrane / protein monoubiquitination / RING-type E3 ubiquitin transferase / ubiquitin-protein transferase activity / transferase activity / protein ubiquitination / zinc ion binding Similarity search - Function | |||||||||
| Biological species |  Thermothelomyces thermophilus ATCC 42464 (fungus) / synthetic construct (others) Thermothelomyces thermophilus ATCC 42464 (fungus) / synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Peiqiang F / Tom R | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: A peroxisomal ubiquitin ligase complex forms a retrotranslocation channel. Authors: Peiqiang Feng / Xudong Wu / Satchal K Erramilli / Joao A Paulo / Pawel Knejski / Steven P Gygi / Anthony A Kossiakoff / Tom A Rapoport /  Abstract: Peroxisomes are ubiquitous organelles that house various metabolic reactions and are essential for human health. Luminal peroxisomal proteins are imported from the cytosol by mobile receptors, which ...Peroxisomes are ubiquitous organelles that house various metabolic reactions and are essential for human health. Luminal peroxisomal proteins are imported from the cytosol by mobile receptors, which then recycle back to the cytosol by a poorly understood process. Recycling requires receptor modification by a membrane-embedded ubiquitin ligase complex comprising three RING finger domain-containing proteins (Pex2, Pex10 and Pex12). Here we report a cryo-electron microscopy structure of the ligase complex, which together with biochemical and in vivo experiments reveals its function as a retrotranslocation channel for peroxisomal import receptors. Each subunit of the complex contributes five transmembrane segments that co-assemble into an open channel. The three ring finger domains form a cytosolic tower, with ring finger 2 (RF2) positioned above the channel pore. We propose that the N terminus of a recycling receptor is inserted from the peroxisomal lumen into the pore and monoubiquitylated by RF2 to enable extraction into the cytosol. If recycling is compromised, receptors are polyubiquitylated by the concerted action of RF10 and RF12 and degraded. This polyubiquitylation pathway also maintains the homeostasis of other peroxisomal import factors. Our results clarify a crucial step during peroxisomal protein import and reveal why mutations in the ligase complex cause human disease. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25750.map.gz emd_25750.map.gz | 62.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25750-v30.xml emd-25750-v30.xml emd-25750.xml emd-25750.xml | 17.4 KB 17.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_25750.png emd_25750.png | 55.8 KB | ||
| Filedesc metadata |  emd-25750.cif.gz emd-25750.cif.gz | 6.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25750 http://ftp.pdbj.org/pub/emdb/structures/EMD-25750 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25750 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25750 | HTTPS FTP |
-Related structure data
| Related structure data |  7t92MC  7t9xC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25750.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25750.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of the peroxisomal retro-translocon formed by a heterotrimeric ubiquitin ligase complex | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
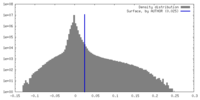
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
+Entire : Peroxisomal heterotrimeric ubiquitin ligase complex bound with Fab
+Supramolecule #1: Peroxisomal heterotrimeric ubiquitin ligase complex bound with Fab
+Supramolecule #2: Peroxin12, Peroxin2, Peroxin10
+Supramolecule #3: Fab heavy chain, Fab light chain
+Macromolecule #1: Peroxin-12
+Macromolecule #2: Peroxin-2
+Macromolecule #3: Peroxin-10
+Macromolecule #4: Fab heavy chain
+Macromolecule #5: Fab light chain
+Macromolecule #6: ZINC ION
+Macromolecule #7: 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
+Macromolecule #8: CHOLESTEROL
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 4 K / Instrument: FEI VITROBOT MARK I |
| Details | This sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.1 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 121644 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)