+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7517 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Rhodopsin-Gi | |||||||||||||||
 Map data Map data | primary map | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Rhodopsin / G protein / cryo-EM / Structure / SIGNALING PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationOpsins / rod bipolar cell differentiation / sperm head plasma membrane / absorption of visible light / G protein-coupled opsin signaling pathway / 11-cis retinal binding / podosome assembly / G protein-coupled photoreceptor activity / photoreceptor inner segment membrane / cellular response to light stimulus ...Opsins / rod bipolar cell differentiation / sperm head plasma membrane / absorption of visible light / G protein-coupled opsin signaling pathway / 11-cis retinal binding / podosome assembly / G protein-coupled photoreceptor activity / photoreceptor inner segment membrane / cellular response to light stimulus / VxPx cargo-targeting to cilium / rod photoreceptor outer segment / thermotaxis / Golgi-associated vesicle membrane / detection of temperature stimulus involved in thermoception / response to light intensity / photoreceptor cell maintenance / G-protein activation / Activation of the phototransduction cascade / Glucagon-type ligand receptors / Thromboxane signalling through TP receptor / Sensory perception of sweet, bitter, and umami (glutamate) taste / G beta:gamma signalling through PI3Kgamma / G beta:gamma signalling through CDC42 / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Ca2+ pathway / G alpha (z) signalling events / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Adrenaline,noradrenaline inhibits insulin secretion / ADP signalling through P2Y purinoceptor 12 / G alpha (q) signalling events / G alpha (i) signalling events / Thrombin signalling through proteinase activated receptors (PARs) / Activation of G protein gated Potassium channels / G-protein activation / G beta:gamma signalling through PI3Kgamma / Prostacyclin signalling through prostacyclin receptor / G beta:gamma signalling through PLC beta / ADP signalling through P2Y purinoceptor 1 / Thromboxane signalling through TP receptor / Presynaptic function of Kainate receptors / G beta:gamma signalling through CDC42 / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G alpha (12/13) signalling events / Glucagon-type ligand receptors / G beta:gamma signalling through BTK / ADP signalling through P2Y purinoceptor 12 / Adrenaline,noradrenaline inhibits insulin secretion / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / Ca2+ pathway / Thrombin signalling through proteinase activated receptors (PARs) / G alpha (z) signalling events / Extra-nuclear estrogen signaling / ciliary membrane / G alpha (s) signalling events / photoreceptor outer segment membrane / G alpha (q) signalling events / spectrin binding / G alpha (i) signalling events / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / Vasopressin regulates renal water homeostasis via Aquaporins / The canonical retinoid cycle in rods (twilight vision) / alkylglycerophosphoethanolamine phosphodiesterase activity / phototransduction, visible light / phototransduction / photoreceptor outer segment / adenylate cyclase inhibitor activity / positive regulation of protein localization to cell cortex / T cell migration / Adenylate cyclase inhibitory pathway / response to prostaglandin E / D2 dopamine receptor binding / cardiac muscle cell apoptotic process / photoreceptor inner segment / G protein-coupled serotonin receptor binding / adenylate cyclase regulator activity / adenylate cyclase-inhibiting serotonin receptor signaling pathway / visual perception / cellular response to forskolin / regulation of mitotic spindle organization / Regulation of insulin secretion / positive regulation of cholesterol biosynthetic process / electron transport chain / negative regulation of insulin secretion / G protein-coupled receptor binding / G protein-coupled receptor activity / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / response to peptide hormone / microtubule cytoskeleton organization / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / centriolar satellite / G-protein beta/gamma-subunit complex binding / Activation of the phototransduction cascade / ADP signalling through P2Y purinoceptor 12 / cell-cell junction Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.5 Å | |||||||||||||||
 Authors Authors | Kang Y / Xu HE | |||||||||||||||
| Funding support |  United States, United States,  China, 4 items China, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2018 Journal: Nature / Year: 2018Title: Cryo-EM structure of human rhodopsin bound to an inhibitory G protein. Authors: Yanyong Kang / Oleg Kuybeda / Parker W de Waal / Somnath Mukherjee / Ned Van Eps / Przemyslaw Dutka / X Edward Zhou / Alberto Bartesaghi / Satchal Erramilli / Takefumi Morizumi / Xin Gu / ...Authors: Yanyong Kang / Oleg Kuybeda / Parker W de Waal / Somnath Mukherjee / Ned Van Eps / Przemyslaw Dutka / X Edward Zhou / Alberto Bartesaghi / Satchal Erramilli / Takefumi Morizumi / Xin Gu / Yanting Yin / Ping Liu / Yi Jiang / Xing Meng / Gongpu Zhao / Karsten Melcher / Oliver P Ernst / Anthony A Kossiakoff / Sriram Subramaniam / H Eric Xu /     Abstract: G-protein-coupled receptors comprise the largest family of mammalian transmembrane receptors. They mediate numerous cellular pathways by coupling with downstream signalling transducers, including the ...G-protein-coupled receptors comprise the largest family of mammalian transmembrane receptors. They mediate numerous cellular pathways by coupling with downstream signalling transducers, including the hetrotrimeric G proteins G (stimulatory) and G (inhibitory) and several arrestin proteins. The structural mechanisms that define how G-protein-coupled receptors selectively couple to a specific type of G protein or arrestin remain unknown. Here, using cryo-electron microscopy, we show that the major interactions between activated rhodopsin and G are mediated by the C-terminal helix of the G α-subunit, which is wedged into the cytoplasmic cavity of the transmembrane helix bundle and directly contacts the amino terminus of helix 8 of rhodopsin. Structural comparisons of inactive, G-bound and arrestin-bound forms of rhodopsin with inactive and G-bound forms of the β-adrenergic receptor provide a foundation to understand the unique structural signatures that are associated with the recognition of G, G and arrestin by activated G-protein-coupled receptors. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7517.map.gz emd_7517.map.gz | 5.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7517-v30.xml emd-7517-v30.xml emd-7517.xml emd-7517.xml | 26.5 KB 26.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7517.png emd_7517.png | 47.3 KB | ||
| Filedesc metadata |  emd-7517.cif.gz emd-7517.cif.gz | 8.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7517 http://ftp.pdbj.org/pub/emdb/structures/EMD-7517 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7517 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7517 | HTTPS FTP |
-Related structure data
| Related structure data |  6cmoMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7517.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7517.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | primary map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.088 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
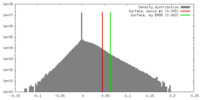
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Protein complex of Rhodopsin with Galphai
| Entire | Name: Protein complex of Rhodopsin with Galphai |
|---|---|
| Components |
|
-Supramolecule #1: Protein complex of Rhodopsin with Galphai
| Supramolecule | Name: Protein complex of Rhodopsin with Galphai / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 180 kDa/nm |
-Macromolecule #1: chimera protein of Soluble cytochrome b562 and Rhodopsin
| Macromolecule | Name: chimera protein of Soluble cytochrome b562 and Rhodopsin type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 52.385406 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: FADYKDDDDA KLQTMHHHHH HHHHHENLYF QGGTADLEDN WETLNDNLKV IEKADNAAQV KDALTKMRAA ALDAQKATPP KLEDKSPDS PEMKDFRHGF DILVGQIDDA LKLANEGKVK EAQAAAEQLK TTRNAYIQKY LMCGTEGPNF YVPFSNATGV V RSPFEYPQ ...String: FADYKDDDDA KLQTMHHHHH HHHHHENLYF QGGTADLEDN WETLNDNLKV IEKADNAAQV KDALTKMRAA ALDAQKATPP KLEDKSPDS PEMKDFRHGF DILVGQIDDA LKLANEGKVK EAQAAAEQLK TTRNAYIQKY LMCGTEGPNF YVPFSNATGV V RSPFEYPQ YYLAEPWQFS MLAAYMFLLI VLGFPINFLT LYVTVQHKKL RTPLNYILLN LAVADLFMVL GGFTSTLYTS LH GYFVFGP TGCNLQGFFA TLGGEIALWS LVVLAIERYV VVCKPMSNFR FGENHAIMGV AFTWVMALAC AAPPLAGWSR YIP EGLQCS CGIDYYTLKP EVNNESFVIY MFVVHFTIPM IIIFFCYGQL VFTVKEAAAQ QQESATTQKA EKEVTRMVII YVIA FLICW VPYASVAFYI FTHQGSCFGP IFMTIPAFFA KSAAIYNPVI YIMMNKQFRN CMLTTICC UniProtKB: Soluble cytochrome b562, Rhodopsin |
-Macromolecule #2: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.445059 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTTGIVETH FTFKDLHFKM FDVGAQRSER KKWIHCFEGV TAIIFCVALS DYDLVLAEDE EM NRMHESM KLFDSICNNK WFTDTSIILF LNKKDLFEEK IKKSPLTICY PEYAGSNTYE EAAAYIQCQF EDLNKRKDTK EIY THFTCS TDTKNVQFVF DAVTDVIIKN NLKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 37.915496 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSLLQSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSASQ DGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG YLSCCRFLDD N QIVTSSGD ...String: MGSLLQSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSASQ DGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG YLSCCRFLDD N QIVTSSGD TTCALWDIET GQQTTTFTGH TGDVMSLSLA PDTRLFVSGA CDASAKLWDV REGMCRQTFT GHESDINAIC FF PNGNAFA TGSDDATCRL FDLRADQELM TYSHDNIICG ITSVSFSKSG RLLLAGYDDF NCNVWDALKA DRAGVLAGHD NRV SCLGVT DDGMAVATGS WDSFLKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 7.56375 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFC UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: Fab light chain
| Macromolecule | Name: Fab light chain / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 23.258783 KDa |
| Recombinant expression | Organism:  Bacteria Latreille et al. 1825 (Bacteria stick insect) Bacteria Latreille et al. 1825 (Bacteria stick insect) |
| Sequence | String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQSSSSLITF GQGTKVEIKR TVAAPSVFIF PPSDSQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD ...String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQSSSSLITF GQGTKVEIKR TVAAPSVFIF PPSDSQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD SKDSTYSLSS TLTLSKADYE KHKVYACEVT HQGLSSPVTK SFNRGEC |
-Macromolecule #6: Fab Heavy chain
| Macromolecule | Name: Fab Heavy chain / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 25.788822 KDa |
| Recombinant expression | Organism:  Bacteria Latreille et al. 1825 (Bacteria stick insect) Bacteria Latreille et al. 1825 (Bacteria stick insect) |
| Sequence | String: EISEVQLVES GGGLVQPGGS LRLSCAASGF NFYYSSIHWV RQAPGKGLEW VASIYSYSGS TSYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC ARYPWYWWME KPYLSLYGMD YWGQGTLVTV SSASTKGPSV FPLAPSSKST SGGTAALGCL V KDYFPEPV ...String: EISEVQLVES GGGLVQPGGS LRLSCAASGF NFYYSSIHWV RQAPGKGLEW VASIYSYSGS TSYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC ARYPWYWWME KPYLSLYGMD YWGQGTLVTV SSASTKGPSV FPLAPSSKST SGGTAALGCL V KDYFPEPV TVSWNSGALT SGVHTFPAVL QSSGLYSLSS VVTVPSSSLG TQTYICNVNH KPSNTKVDKK VEPKSCDKTH T |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 9.0 mg/mL |
|---|---|
| Buffer | pH: 7.2 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 1.92 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)