[English] 日本語
 Yorodumi
Yorodumi- EMDB-41915: Cryo-EM structure of human claudin-4 complex with Clostridium per... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human claudin-4 complex with Clostridium perfringens enterotoxin C-terminal domain and sFab COP-1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Claudin / Fab / Toxin / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationparacellular transport / calcium-independent cell-cell adhesion / Tight junction interactions / bicellular tight junction assembly / apicolateral plasma membrane / Mo-molybdopterin cofactor biosynthetic process / regulation of cell morphogenesis / tight junction / positive regulation of wound healing / renal absorption ...paracellular transport / calcium-independent cell-cell adhesion / Tight junction interactions / bicellular tight junction assembly / apicolateral plasma membrane / Mo-molybdopterin cofactor biosynthetic process / regulation of cell morphogenesis / tight junction / positive regulation of wound healing / renal absorption / chloride channel activity / establishment of skin barrier / lateral plasma membrane / bicellular tight junction / chloride channel complex / response to progesterone / basal plasma membrane / female pregnancy / circadian rhythm / cell-cell junction / transmembrane signaling receptor activity / cell adhesion / apical plasma membrane / positive regulation of cell migration / structural molecule activity / identical protein binding / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 5.3 Å | |||||||||
 Authors Authors | Vecchio AJ | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2024 Journal: Commun Biol / Year: 2024Title: Structural and biophysical insights into targeting of claudin-4 by a synthetic antibody fragment. Authors: Satchal K Erramilli / Pawel K Dominik / Chinemerem P Ogbu / Anthony A Kossiakoff / Alex J Vecchio /  Abstract: Claudins are a 27-member family of ~25 kDa membrane proteins that integrate into tight junctions to form molecular barriers at the paracellular spaces between endothelial and epithelial cells. As ...Claudins are a 27-member family of ~25 kDa membrane proteins that integrate into tight junctions to form molecular barriers at the paracellular spaces between endothelial and epithelial cells. As the backbone of tight junction structure and function, claudins are attractive targets for modulating tissue permeability to deliver drugs or treat disease. However, structures of claudins are limited due to their small sizes and physicochemical properties-these traits also make therapy development a challenge. Here we report the development of a synthetic antibody fragment (sFab) that binds human claudin-4 and the determination of a high-resolution structure of it bound to claudin-4/enterotoxin complexes using cryogenic electron microscopy. Structural and biophysical results reveal this sFabs mechanism of select binding to human claudin-4 over other homologous claudins and establish the ability of sFabs to bind hard-to-target claudins to probe tight junction structure and function. The findings provide a framework for tight junction modulation by sFabs for tissue-selective therapies. #1:  Journal: bioRxiv / Year: 2023 Journal: bioRxiv / Year: 2023Title: Cryo-EM structures of a synthetic antibody against 22 kDa claudin-4 reveal its complex with Authors: Erramilli SK / Dominik PK / Ogbu CP / Kossiakoff AA / Vecchio AJ | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41915.map.gz emd_41915.map.gz | 250.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41915-v30.xml emd-41915-v30.xml emd-41915.xml emd-41915.xml | 22.6 KB 22.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41915_fsc.xml emd_41915_fsc.xml | 23.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_41915.png emd_41915.png | 33.6 KB | ||
| Masks |  emd_41915_msk_1.map emd_41915_msk_1.map | 512 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-41915.cif.gz emd-41915.cif.gz | 7.1 KB | ||
| Others |  emd_41915_half_map_1.map.gz emd_41915_half_map_1.map.gz emd_41915_half_map_2.map.gz emd_41915_half_map_2.map.gz | 474.5 MB 474.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41915 http://ftp.pdbj.org/pub/emdb/structures/EMD-41915 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41915 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41915 | HTTPS FTP |
-Related structure data
| Related structure data |  8u5bMC  8u4vC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41915.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41915.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.507 Å | ||||||||||||||||||||||||||||||||||||
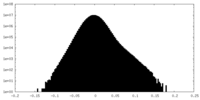
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_41915_msk_1.map emd_41915_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
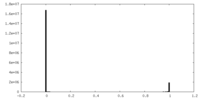
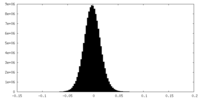
| Density Histograms |
-Half map: #2
| File | emd_41915_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
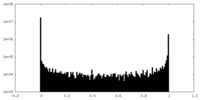
| Density Histograms |
-Half map: #1
| File | emd_41915_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
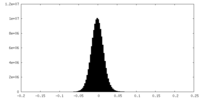
| Density Histograms |
- Sample components
Sample components
-Entire : Human claudin-4 complex with Clostridium perfringens enterotoxin ...
| Entire | Name: Human claudin-4 complex with Clostridium perfringens enterotoxin C-terminal domain and sFab COP-1 |
|---|---|
| Components |
|
-Supramolecule #1: Human claudin-4 complex with Clostridium perfringens enterotoxin ...
| Supramolecule | Name: Human claudin-4 complex with Clostridium perfringens enterotoxin C-terminal domain and sFab COP-1 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 Details: Assembled complex of 4 proteins (Fab is 2 proteins) expressed from insect cells and E coli |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 100 KDa |
-Macromolecule #1: Claudin-4
| Macromolecule | Name: Claudin-4 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.613852 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MASMGLQVMG IALAVLGWLA VMLCCALPMW RVTAFIGSNI VTSQTIWEGL WMNCVVQSTG QMQCKVYDSL LALPQDLQAA RALVIISII VAALGVLLSV VGGKCTNCLE DESAKAKTMI VAGVVFLLAG LMVIVPVSWT AHNIIQDFYN PLVASGQKRE M GASLYVGW ...String: MASMGLQVMG IALAVLGWLA VMLCCALPMW RVTAFIGSNI VTSQTIWEGL WMNCVVQSTG QMQCKVYDSL LALPQDLQAA RALVIISII VAALGVLLSV VGGKCTNCLE DESAKAKTMI VAGVVFLLAG LMVIVPVSWT AHNIIQDFYN PLVASGQKRE M GASLYVGW AASGLLLLGG GLLCCNCPPR TDKPYSAKYS AARSAAASNY VGLVPR UniProtKB: Claudin-4 |
-Macromolecule #2: Heat-labile enterotoxin B chain
| Macromolecule | Name: Heat-labile enterotoxin B chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.044594 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: GSDEILDLAA ATERLNLTDA LNSNPAGNLY DWRSSNSYPW TQKLNLHLTI TATGQKYRIL ASKIVDFNIY SNNFNNLVKL EQSLGDGVK DHYVDISLDA GQYVLVMKAN SSYSGNYPYS ILFQKF UniProtKB: Molybdopterin biosynthesis protein MoaB |
-Macromolecule #3: COP-1 sFab Light Chain
| Macromolecule | Name: COP-1 sFab Light Chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 23.258783 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQSSSSLITF GQGTKVEIKR TVAAPSVFIF PPSDSQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD ...String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQSSSSLITF GQGTKVEIKR TVAAPSVFIF PPSDSQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD SKDSTYSLSS TLTLSKADYE KHKVYACEVT HQGLSSPVTK SFNRGEC |
-Macromolecule #4: COP-1 sFab Heavy Chain
| Macromolecule | Name: COP-1 sFab Heavy Chain / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 28.064498 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKKNIAFLLA SMFVFSIATN AYAEISEVQL VESGGGLVQP GGSLRLSCAA SGFNFSSSYI HWVRQAPGKG LEWVASISSS SGSTSYADS VKGRFTISAD TSKNTAYLQM NSLRAEDTAV YYCARWFHPW WWWEYLFRGA IDYWGQGTLV TVSSASTKGP S VFPLAPSS ...String: MKKNIAFLLA SMFVFSIATN AYAEISEVQL VESGGGLVQP GGSLRLSCAA SGFNFSSSYI HWVRQAPGKG LEWVASISSS SGSTSYADS VKGRFTISAD TSKNTAYLQM NSLRAEDTAV YYCARWFHPW WWWEYLFRGA IDYWGQGTLV TVSSASTKGP S VFPLAPSS KSTSGGTAAL GCLVKDYFPE PVTVSWNSGA LTSGVHTFPA VLQSSGLYSL SSVVTVPSSS LGTQTYICNV NH KPSNTKV DKKVEPKSCD KTHT |
-Macromolecule #5: Lauryl Maltose Neopentyl Glycol
| Macromolecule | Name: Lauryl Maltose Neopentyl Glycol / type: ligand / ID: 5 / Number of copies: 1 / Formula: AV0 |
|---|---|
| Molecular weight | Theoretical: 1.005188 KDa |
| Chemical component information |  ChemComp-AV0: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 20 mM Hepes pH 7.4, 100 mM NaCl, and 0.003% LMNG |
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 1073 / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 92000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)