[English] 日本語
 Yorodumi
Yorodumi- EMDB-20806: Single-Particle Cryo-EM Structure of Plasmodium falciparum Chloro... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20806 | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Single-Particle Cryo-EM Structure of Plasmodium falciparum Chloroquine Resistance Transporter (PfCRT) 7G8 Isoform | ||||||||||||||||||||||||||||||
 Map data Map data | Sharpened map | ||||||||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||||||||
 Keywords Keywords | Transporter / drug resistance / digestive vacuole of malaria parasite / nanodisc / MEMBRANE PROTEIN | ||||||||||||||||||||||||||||||
| Function / homology | Chloroquine-resistance transporter-like / Chloroquine resistance transporter / CRT-like, chloroquine-resistance transporter-like / vacuolar membrane / amino acid transport / xenobiotic transmembrane transporter activity / iron ion transport / Chloroquine resistance transporter Function and homology information Function and homology information | ||||||||||||||||||||||||||||||
| Biological species |   Homo sapiens (human) / Homo sapiens (human) /  | ||||||||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | ||||||||||||||||||||||||||||||
 Authors Authors | Kim J / Tan YZ | ||||||||||||||||||||||||||||||
| Funding support |  United States, 9 items United States, 9 items
| ||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2019 Journal: Nature / Year: 2019Title: Structure and drug resistance of the Plasmodium falciparum transporter PfCRT. Authors: Jonathan Kim / Yong Zi Tan / Kathryn J Wicht / Satchal K Erramilli / Satish K Dhingra / John Okombo / Jeremie Vendome / Laura M Hagenah / Sabrina I Giacometti / Audrey L Warren / Kamil Nosol ...Authors: Jonathan Kim / Yong Zi Tan / Kathryn J Wicht / Satchal K Erramilli / Satish K Dhingra / John Okombo / Jeremie Vendome / Laura M Hagenah / Sabrina I Giacometti / Audrey L Warren / Kamil Nosol / Paul D Roepe / Clinton S Potter / Bridget Carragher / Anthony A Kossiakoff / Matthias Quick / David A Fidock / Filippo Mancia /  Abstract: The emergence and spread of drug-resistant Plasmodium falciparum impedes global efforts to control and eliminate malaria. For decades, treatment of malaria has relied on chloroquine (CQ), a safe and ...The emergence and spread of drug-resistant Plasmodium falciparum impedes global efforts to control and eliminate malaria. For decades, treatment of malaria has relied on chloroquine (CQ), a safe and affordable 4-aminoquinoline that was highly effective against intra-erythrocytic asexual blood-stage parasites, until resistance arose in Southeast Asia and South America and spread worldwide. Clinical resistance to the chemically related current first-line combination drug piperaquine (PPQ) has now emerged regionally, reducing its efficacy. Resistance to CQ and PPQ has been associated with distinct sets of point mutations in the P. falciparum CQ-resistance transporter PfCRT, a 49-kDa member of the drug/metabolite transporter superfamily that traverses the membrane of the acidic digestive vacuole of the parasite. Here we present the structure, at 3.2 Å resolution, of the PfCRT isoform of CQ-resistant, PPQ-sensitive South American 7G8 parasites, using single-particle cryo-electron microscopy and antigen-binding fragment technology. Mutations that contribute to CQ and PPQ resistance localize primarily to moderately conserved sites on distinct helices that line a central negatively charged cavity, indicating that this cavity is the principal site of interaction with the positively charged CQ and PPQ. Binding and transport studies reveal that the 7G8 isoform binds both drugs with comparable affinities, and that these drugs are mutually competitive. The 7G8 isoform transports CQ in a membrane potential- and pH-dependent manner, consistent with an active efflux mechanism that drives CQ resistance, but does not transport PPQ. Functional studies on the newly emerging PfCRT F145I and C350R mutations, associated with decreased PPQ susceptibility in Asia and South America, respectively, reveal their ability to mediate PPQ transport in 7G8 variant proteins and to confer resistance in gene-edited parasites. Structural, functional and in silico analyses suggest that distinct mechanistic features mediate the resistance to CQ and PPQ in PfCRT variants. These data provide atomic-level insights into the molecular mechanism of this key mediator of antimalarial treatment failures. | ||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20806.map.gz emd_20806.map.gz | 95.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20806-v30.xml emd-20806-v30.xml emd-20806.xml emd-20806.xml | 44.1 KB 44.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20806_fsc_1.xml emd_20806_fsc_1.xml emd_20806_fsc_2.xml emd_20806_fsc_2.xml | 11.4 KB 11.4 KB | Display Display |  FSC data file FSC data file |
| Images |  emd_20806.png emd_20806.png | 239.2 KB | ||
| Masks |  emd_20806_msk_1.map emd_20806_msk_1.map emd_20806_msk_2.map emd_20806_msk_2.map | 103 MB 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-20806.cif.gz emd-20806.cif.gz | 7.9 KB | ||
| Others |  emd_20806_additional_1.map.gz emd_20806_additional_1.map.gz emd_20806_additional_2.map.gz emd_20806_additional_2.map.gz emd_20806_additional_3.map.gz emd_20806_additional_3.map.gz emd_20806_additional_4.map.gz emd_20806_additional_4.map.gz emd_20806_additional_5.map.gz emd_20806_additional_5.map.gz emd_20806_additional_6.map.gz emd_20806_additional_6.map.gz emd_20806_additional_7.map.gz emd_20806_additional_7.map.gz emd_20806_additional_8.map.gz emd_20806_additional_8.map.gz emd_20806_half_map_1.map.gz emd_20806_half_map_1.map.gz emd_20806_half_map_2.map.gz emd_20806_half_map_2.map.gz | 95.5 MB 566.8 KB 13.9 MB 12.1 MB 173.1 KB 14.9 MB 12.6 MB 172.8 KB 15.9 MB 15.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20806 http://ftp.pdbj.org/pub/emdb/structures/EMD-20806 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20806 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20806 | HTTPS FTP |
-Related structure data
| Related structure data |  6ukjMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10330 (Title: 3.2 Å Single-Particle Cryo-EM Reconstruction of 49 kDa Membrane-Bound PfCRT Complexed with Fab EMPIAR-10330 (Title: 3.2 Å Single-Particle Cryo-EM Reconstruction of 49 kDa Membrane-Bound PfCRT Complexed with FabData size: 830.4 Data #1: Unaligned and compressed multi-frame micrographs [micrographs - multiframe] Data #2: Aligned and magnification anisotropy corrected micrographs [micrographs - single frame] Data #3: Final Particle Stack with Refined Euler Angles and Shifts [picked particles - single frame - processed]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_20806.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20806.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.035 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
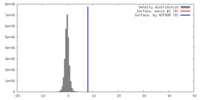
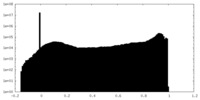
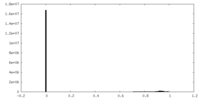
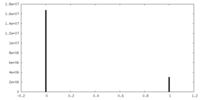
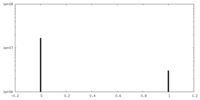
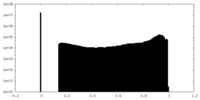
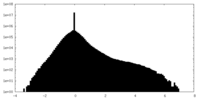
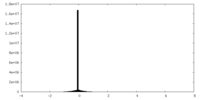
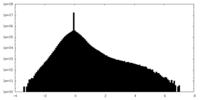
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
+Mask #1
+Mask #2
+Additional map: Unsharpened map
+Additional map: Local resolution map
+Additional map: 3DFSC - PfCRT with Fab, Raw
+Additional map: 3DFSC - PfCRT with Fab, Thresholded
+Additional map: 3DFSC - PfCRT with Fab, Thresholded and Binarized
+Additional map: 3DFSC - PfCRT only, Raw
+Additional map: 3DFSC - PfCRT Only, Thresholded
+Additional map: 3DFSC - PfCRT only, Thresholded and binarized
+Half map: Half map 1
+Half map: Half map 2
- Sample components
Sample components
-Entire : Plasmodium falciparum Chloroquine Resistance Transporter (PfCRT) ...
| Entire | Name: Plasmodium falciparum Chloroquine Resistance Transporter (PfCRT) 7G8 Isoform Complexed with Fab |
|---|---|
| Components |
|
-Supramolecule #1: Plasmodium falciparum Chloroquine Resistance Transporter (PfCRT) ...
| Supramolecule | Name: Plasmodium falciparum Chloroquine Resistance Transporter (PfCRT) 7G8 Isoform Complexed with Fab type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Molecular weight | Theoretical: 102 KDa |
-Supramolecule #2: Plasmodium falciparum Chloroquine Resistance Transporter (PfCRT)
| Supramolecule | Name: Plasmodium falciparum Chloroquine Resistance Transporter (PfCRT) type: organelle_or_cellular_component / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 49 KDa |
-Supramolecule #3: Antigen-binding fragment (Fab)
| Supramolecule | Name: Antigen-binding fragment (Fab) / type: organelle_or_cellular_component / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 50 KDa |
-Macromolecule #1: Chloroquine resistance transporter
| Macromolecule | Name: Chloroquine resistance transporter / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 53.074461 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGRAKFASKK NNQKNSSKND ERYRELDNLV QEGNGSRLGG GSCLGKCAHV FKLIFKEIKD NIFIYILSII YLSVSVMNTI FAKRTLNKI GNYSFVTSET HNFICMIMFF IVYSLFGNKK GNSKERHRSF NLQFFAISML DACSVILAFI GLTRTTGNIQ S FVLQLSIP ...String: MGRAKFASKK NNQKNSSKND ERYRELDNLV QEGNGSRLGG GSCLGKCAHV FKLIFKEIKD NIFIYILSII YLSVSVMNTI FAKRTLNKI GNYSFVTSET HNFICMIMFF IVYSLFGNKK GNSKERHRSF NLQFFAISML DACSVILAFI GLTRTTGNIQ S FVLQLSIP INMFFCFLIL RYRYHLYNYL GAVIIVVTIA LVEMKLSFET QEENSIIFNL VLISSLIPVC FSNMTREIVF KK YKIDILR LNAMVSFFQL FTSCLILPVY TLPFLKQLHL PYNEIWTNIK NGFACLFLGR NTVVENCGLG MAKLCDDCDG AWK TFALFS FFDICDNLIT SYIIDKFSTM TYTIVSCIQG PALAIAYYFK FLAGDVVREP RLLDFVTLFG YLFGSIIYRV GNII LERKK MRNEENEDSE GELTNVDSII TQAAAGGGSG GGSENLYFQG SHHHHHHHHH HWSHPQFEK UniProtKB: Chloroquine resistance transporter |
-Macromolecule #2: Fab Heavy Chain
| Macromolecule | Name: Fab Heavy Chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.737631 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EISEVQLVES GGGLVQPGGS LRLSCAASGF NVSYSYIHWV RQAPGKGLEW VASIYPYSGY TSYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC ARYGSNYSFW YRGSSVTYAI DYWGQGTLVT VSSASTKGPS VFPLAPSSKS TSGGTAALGC L VKDYFPEP ...String: EISEVQLVES GGGLVQPGGS LRLSCAASGF NVSYSYIHWV RQAPGKGLEW VASIYPYSGY TSYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC ARYGSNYSFW YRGSSVTYAI DYWGQGTLVT VSSASTKGPS VFPLAPSSKS TSGGTAALGC L VKDYFPEP VTVSWNSGAL TSGVHTFPAV LQSSGLYSLS SVVTVPSSSL GTQTYICNVN HKPSNTKVDK KVEPKSCDKT HT C |
-Macromolecule #3: Fab Light Chain
| Macromolecule | Name: Fab Light Chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.355898 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQSSTWPITF GQGTKVEIKR TVAAPSVFIF PPSDSQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD ...String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQSSTWPITF GQGTKVEIKR TVAAPSVFIF PPSDSQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD SKDSTYSLSS TLTLSKADYE KHKVYACEVT HQGLSSPVTK SFNRGEC |
-Macromolecule #4: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 4 / Number of copies: 1 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.56 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7 Component:
Details: Solution was filtered and degassed. | |||||||||
| Grid | Model: UltrAuFoil / Material: GOLD / Mesh: 300 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 7 sec. / Pretreatment - Atmosphere: OTHER | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 279 K / Instrument: LEICA EM GP / Details: Blot for 2 seconds before plunging.. | |||||||||
| Details | Protein was incorporated into lipid nanodiscs and complexed with Fab. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Spherical aberration corrector: Cs corrector was used. / Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-80 / Number grids imaged: 1 / Number real images: 3377 / Average exposure time: 6.0 sec. / Average electron dose: 91.56 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.001 mm / Nominal defocus max: 0.0018000000000000002 µm / Nominal defocus min: 0.0012 µm / Nominal magnification: 215000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Details | Rosetta was used to generate an ab initio model for the PfCRT using the map. 4XMM chains H and L were used as starting point to model the Fab. | ||||||
| Refinement | Space: REAL / Protocol: AB INITIO MODEL | ||||||
| Output model |  PDB-6ukj: |
 Movie
Movie Controller
Controller













 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)