[English] 日本語
 Yorodumi
Yorodumi- EMDB-40623: Chlorella virus Hyaluronan Synthase bound to GlcNAc primer and UD... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Chlorella virus Hyaluronan Synthase bound to GlcNAc primer and UDP-GlcA | |||||||||
 Map data Map data | Chlorella virus Hyaluronan Synthase bound to GlcNAc primer and UDP-GlcA | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | hyaluronic acid / hyaluronan / HA / HAS / glycosyltransferase / GT / membrane protein / nanobody / n-acetylglucosamine / glucuronic acid / TRANSFERASE | |||||||||
| Function / homology | hyaluronan synthase activity / Glycosyltransferase like family 2 / hyaluronan biosynthetic process / Nucleotide-diphospho-sugar transferases / plasma membrane / Hyaluronan synthase Function and homology information Function and homology information | |||||||||
| Biological species |   Paramecium bursaria Chlorella virus 1 / Paramecium bursaria Chlorella virus 1 /  Paramecium bursaria Chlorella virus CZ-2 / Paramecium bursaria Chlorella virus CZ-2 /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Stephens Z / Zimmer J | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2025 Journal: Nat Struct Mol Biol / Year: 2025Title: Structural insights into translocation and tailored synthesis of hyaluronan. Authors: Ireneusz Górniak / Zachery Stephens / Satchal K Erramilli / Tomasz Gawda / Anthony A Kossiakoff / Jochen Zimmer /  Abstract: Hyaluronan (HA) is an essential component of the vertebrate extracellular matrix. It is a heteropolysaccharide of N-acetylglucosamine (GlcNAc) and glucuronic acid (GlcA) reaching several megadaltons ...Hyaluronan (HA) is an essential component of the vertebrate extracellular matrix. It is a heteropolysaccharide of N-acetylglucosamine (GlcNAc) and glucuronic acid (GlcA) reaching several megadaltons in healthy tissues. HA is synthesized and translocated in a coupled reaction by HA synthase (HAS). Here, structural snapshots of HAS provide insights into HA biosynthesis, from substrate recognition to HA elongation and translocation. We monitor the extension of a GlcNAc primer with GlcA, reveal the coordination of the uridine diphosphate product by a conserved gating loop and capture the opening of a translocation channel to coordinate a translocating HA polymer. Furthermore, we identify channel-lining residues that modulate HA product lengths. Integrating structural and biochemical analyses suggests an avenue for polysaccharide engineering based on finely tuned enzymatic activity and HA coordination. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40623.map.gz emd_40623.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40623-v30.xml emd-40623-v30.xml emd-40623.xml emd-40623.xml | 19 KB 19 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_40623.png emd_40623.png | 35.2 KB | ||
| Filedesc metadata |  emd-40623.cif.gz emd-40623.cif.gz | 7 KB | ||
| Others |  emd_40623_half_map_1.map.gz emd_40623_half_map_1.map.gz emd_40623_half_map_2.map.gz emd_40623_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40623 http://ftp.pdbj.org/pub/emdb/structures/EMD-40623 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40623 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40623 | HTTPS FTP |
-Related structure data
| Related structure data |  8sndMC  8smmC  8smnC  8smpC  8sncC  8sneC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_40623.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40623.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Chlorella virus Hyaluronan Synthase bound to GlcNAc primer and UDP-GlcA | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
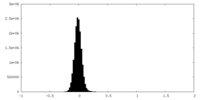
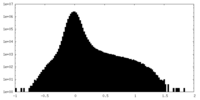
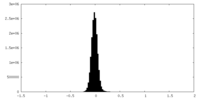
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half Map B
| File | emd_40623_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map A
| File | emd_40623_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Ternary complex of Chlorella virus hyaluronan synthase bound to n...
+Supramolecule #1: Ternary complex of Chlorella virus hyaluronan synthase bound to n...
+Macromolecule #1: Hyaluronan synthase
+Macromolecule #2: Nanobody 872
+Macromolecule #3: Nanobody 881
+Macromolecule #4: 1,2-Distearoyl-sn-glycerophosphoethanolamine
+Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
+Macromolecule #6: URIDINE-5'-DIPHOSPHATE-GLUCURONIC ACID
+Macromolecule #7: CHOLESTEROL HEMISUCCINATE
+Macromolecule #8: MANGANESE (II) ION
+Macromolecule #9: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 81000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.1 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 176543 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)