-Search query
-Search result
Showing 1 - 50 of 134 items for (author: cook & ad)

EMDB-53311: 
Cryo-EM map of SKM-70S ribosomal stalled complex in the major state (vacant A-site, canon)
Method: single particle / : Morici M, Corazza M, Safdari HA, Wilson DN

EMDB-53341: 
Cryo-EM structure of SKM-70S ribosomal stalled complex in the A-tRNA positioned (Body open) state.
Method: single particle / : Morici M, Corazza M, Safdari HA, Wilson DN

EMDB-55145: 
Cryo-EM structure of SKM-70S ribosomal stalled complex in the rotated state with hybrid tRNAs
Method: single particle / : Morici M, Corazza M, Safdari HA, Wilson DN

PDB-9qqq: 
Cryo-EM structure of SKM-70S ribosomal stalled complex in the major state (vacant A-site, canon)
Method: single particle / : Morici M, Corazza M, Safdari HA, Wilson DN

PDB-9qsj: 
Cryo-EM structure of SKM-70S ribosomal stalled complex in the A-tRNA positioned (Body open) state.
Method: single particle / : Morici M, Corazza M, Safdari HA, Wilson DN

PDB-9sro: 
Cryo-EM structure of SKM-70S ribosomal stalled complex in the rotated state with hybrid tRNAs
Method: single particle / : Morici M, Corazza M, Safdari HA, Wilson DN

EMDB-70618: 
Cryo-EM structure of the C. neoformans lipid flippase Apt1-Cdc50 bound with butyrolactol A in the E2P state
Method: single particle / : Duan HD, Li H

PDB-9omv: 
Cryo-EM structure of the C. neoformans lipid flippase Apt1-Cdc50 bound with butyrolactol A in the E2P state
Method: single particle / : Duan HD, Li H

EMDB-48323: 
Structure of the IFIT2-IFIT3 heterodimer from Mus musculus
Method: single particle / : Glasner DR, Todd C, Cook BD, DUrso A, Khosla S, Estrada E, Wagner J, Bartels MD, Ford P, Prych J, Hatch K, Yee BA, Ego KM, Liang Q, Holland SR, Case JB, Corbett KD, Diamond MS, Yeo GW, Herzik Jr MA, Van Nostrand EL, Daugherty MD

PDB-9mk9: 
Structure of the IFIT2-IFIT3 heterodimer from Mus musculus
Method: single particle / : Glasner DR, Todd C, Cook BD, DUrso A, Khosla S, Estrada E, Wagner J, Bartels MD, Ford P, Prych J, Hatch K, Yee BA, Ego KM, Liang Q, Holland SR, Case JB, Corbett KD, Diamond MS, Yeo GW, Herzik Jr MA, Van Nostrand EL, Daugherty MD

EMDB-47339: 
Cryo-EM structure of the C. neoformans lipid flippase Apt1-Cdc50 in the E1 state
Method: single particle / : Duan HD, Li H

PDB-9dzv: 
Cryo-EM structure of the C. neoformans lipid flippase Apt1-Cdc50 in the E1 state
Method: single particle / : Duan HD, Li H

EMDB-42954: 
ACAD11 D220A with 4-hydroxyvaleryl-CoA
Method: single particle / : Zhang J, Bartlett A, Rashan E, Yuan P, Pagliarini D

EMDB-42955: 
ACAD11 D753N with 4-phosphovaleryl-CoA
Method: single particle / : Zhang J, Bartlett A, Rashan E, Yuan P, Pagliarini D

PDB-8v3u: 
ACAD11 D220A with 4-hydroxyvaleryl-CoA
Method: single particle / : Zhang J, Bartlett A, Rashan E, Yuan P, Pagliarini D

PDB-8v3v: 
ACAD11 D753N with 4-phosphovaleryl-CoA
Method: single particle / : Zhang J, Bartlett A, Rashan E, Yuan P, Pagliarini D

EMDB-40180: 
MsbA bound to cerastecin C
Method: single particle / : Chen Y, Klein D

EMDB-17209: 
Human Complement C3b in complex with Trypanosoma brucei ISG65.
Method: single particle / : Cook AD, Higgins MK

EMDB-17219: 
Human Complement C3b in complex with Trypanosoma brucei ISG65
Method: single particle / : Cook AD, Higgins MK

EMDB-17220: 
Human Complement C3b in complex with Trypanosoma brucei ISG65
Method: single particle / : Cook AD, Higgins MK

EMDB-17221: 
Human Complement C3b
Method: single particle / : Cook AD, Higgins MK

EMDB-17273: 
Structure of complement C3 bound to Trypanosoma brucei ISG65
Method: single particle / : Cook AD, Higgins MK

EMDB-43320: 
Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein
Method: single particle / : Zhu X, Mannar D, Saville J, Poloni C, Bezeruk A, Tidey K, Ahmed S, Tuttle K, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S

EMDB-43321: 
Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with mouse ACE2 (conformation 2)
Method: single particle / : Zhu X, Mannar D, Saville J, Poloni C, Bezeruk A, Tidey K, Ahmed S, Tuttle K, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S

EMDB-43322: 
Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with mouse ACE2 (conformation 1)
Method: single particle / : Zhu X, Mannar D, Saville J, Poloni C, Bezeruk A, Tidey K, Ahmed S, Tuttle K, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S

EMDB-43323: 
Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with mouse ACE2 (focused refinement of RBD and mouse ACE2)
Method: single particle / : Zhu X, Mannar D, Saville J, Poloni C, Bezeruk A, Tidey K, Ahmed S, Tuttle K, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S

EMDB-43324: 
Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with human ACE2
Method: single particle / : Zhu X, Mannar D, Saville J, Poloni C, Bezeruk A, Tidey K, Ahmed S, Tuttle K, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S

EMDB-43325: 
Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with human ACE2 (focused refinement of RBD and ACE2)
Method: single particle / : Zhu X, Mannar D, Saville J, Poloni C, Bezeruk A, Tidey K, Ahmed S, Tuttle K, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S

EMDB-43326: 
Negative Stain EM Reconstructions of SARS-CoV-2 spike proteins mixed with polyclonal antibodies from donor 4.
Method: single particle / : Mannar D, Zhu X, Saville J, Poloni C, Bezeruk A, Tidey K, Ahmed S, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S

PDB-8vkk: 
Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein
Method: single particle / : Zhu X, Mannar D, Saville J, Poloni C, Bezeruk A, Tidey K, Ahmed S, Tuttle K, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S

PDB-8vkl: 
Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with mouse ACE2 (conformation 2)
Method: single particle / : Zhu X, Mannar D, Saville J, Poloni C, Bezeruk A, Tidey K, Ahmed S, Tuttle K, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S

PDB-8vkm: 
Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with mouse ACE2 (conformation 1)
Method: single particle / : Zhu X, Mannar D, Saville J, Poloni C, Bezeruk A, Tidey K, Ahmed S, Tuttle K, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S

PDB-8vkn: 
Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with mouse ACE2 (focused refinement of RBD and mouse ACE2)
Method: single particle / : Zhu X, Mannar D, Saville J, Poloni C, Bezeruk A, Tidey K, Ahmed S, Tuttle K, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S

PDB-8vko: 
Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with human ACE2
Method: single particle / : Zhu X, Mannar D, Saville J, Poloni C, Bezeruk A, Tidey K, Ahmed S, Tuttle K, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S

PDB-8vkp: 
Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with human ACE2 (focused refinement of RBD and ACE2)
Method: single particle / : Zhu X, Mannar D, Saville J, Poloni C, Bezeruk A, Tidey K, Ahmed S, Tuttle K, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S

EMDB-42124: 
Cryo-EM structure of human STEAP1 in complex with AMG 509 Fab
Method: single particle / : Li F, Bailis JM

PDB-8ucd: 
Cryo-EM structure of human STEAP1 in complex with AMG 509 Fab
Method: single particle / : Li F, Bailis JM, Zhang H

EMDB-26767: 
The 2.19-angstrom CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - Complex minus stalk
Method: single particle / : Grinter R, Venugopal H, Kropp A, Greening C

EMDB-26801: 
The 1.67 Angstrom CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - catalytic dimer (Huc2S2L)
Method: single particle / : Grinter R, Venugopal H, Kropp A, Greening C

EMDB-26802: 
The CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - Full complex focused refinement of stalk
Method: single particle / : Grinter R, Venugopal H, Kropp A, Greening C

EMDB-27661: 
The 1.52 angstrom CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - catalytic dimer (Huc2S2L)
Method: single particle / : Grinter R, Venugopal H, Kropp A, Greening C

PDB-7utd: 
The 2.19-angstrom CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - Complex minus stalk
Method: single particle / : Grinter R, Venugopal H, Kropp A, Greening C

PDB-7uur: 
The 1.67 Angstrom CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - catalytic dimer (Huc2S2L)
Method: single particle / : Grinter R, Venugopal H, Kropp A, Greening C

PDB-7uus: 
The CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - Full complex focused refinement of stalk
Method: single particle / : Grinter R, Venugopal H, Kropp A, Greening C

PDB-8dqv: 
The 1.52 angstrom CryoEM structure of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis - catalytic dimer (Huc2S2L)
Method: single particle / : Grinter R, Venugopal H, Kropp A, Greening C

EMDB-14460: 
P. berghei kinesin-8B motor domain in AMPPNP state bound to tubulin dimer
Method: single particle / : Liu T, Shilliday F, Cook AD, Moores CA

PDB-7z2b: 
P. berghei kinesin-8B motor domain in AMPPNP state bound to tubulin dimer
Method: single particle / : Liu T, Shilliday F, Cook AD, Moores CA
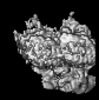
EMDB-14459: 
P. berghei kinesin-8B motor domain in no nucleotide state bound to tubulin dimer
Method: single particle / : Liu T, Shilliday F, Cook AD, Moores CA
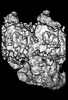
EMDB-14461: 
P. falciparum kinesin-8B motor domain in no nucleotide bound to tubulin dimer
Method: single particle / : Liu T, Shilliday F, Cook AD, Moores CA
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
