[English] 日本語
 Yorodumi
Yorodumi- EMDB-43326: Negative Stain EM Reconstructions of SARS-CoV-2 spike proteins mi... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Negative Stain EM Reconstructions of SARS-CoV-2 spike proteins mixed with polyclonal antibodies from donor 4. | |||||||||
 Map data Map data | Negative Stain EM reconstruction of XBB.1.5 spike protein when mixed with polyclonal fabs from donor 4 (apo - C1 symmetry) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / VIRAL PROTEIN | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 20.0 Å | |||||||||
 Authors Authors | Mannar D / Zhu X / Saville J / Poloni C / Bezeruk A / Tidey K / Ahmed S / Vahdatihassani F / Cholak S / Cook L ...Mannar D / Zhu X / Saville J / Poloni C / Bezeruk A / Tidey K / Ahmed S / Vahdatihassani F / Cholak S / Cook L / Steiner TS / Subramaniam S | |||||||||
| Funding support |  Canada, 1 items Canada, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Altered receptor binding, antibody evasion and retention of T cell recognition by the SARS-CoV-2 XBB.1.5 spike protein. Authors: Dhiraj Mannar / James W Saville / Chad Poloni / Xing Zhu / Alison Bezeruk / Keith Tidey / Sana Ahmed / Katharine S Tuttle / Faezeh Vahdatihassani / Spencer Cholak / Laura Cook / Theodore S ...Authors: Dhiraj Mannar / James W Saville / Chad Poloni / Xing Zhu / Alison Bezeruk / Keith Tidey / Sana Ahmed / Katharine S Tuttle / Faezeh Vahdatihassani / Spencer Cholak / Laura Cook / Theodore S Steiner / Sriram Subramaniam /   Abstract: The XBB.1.5 variant of SARS-CoV-2 has rapidly achieved global dominance and exhibits a high growth advantage over previous variants. Preliminary reports suggest that the success of XBB.1.5 stems from ...The XBB.1.5 variant of SARS-CoV-2 has rapidly achieved global dominance and exhibits a high growth advantage over previous variants. Preliminary reports suggest that the success of XBB.1.5 stems from mutations within its spike glycoprotein, causing immune evasion and enhanced receptor binding. We present receptor binding studies that demonstrate retention of binding contacts with the human ACE2 receptor and a striking decrease in binding to mouse ACE2 due to the revertant R493Q mutation. Despite extensive evasion of antibody binding, we highlight a region on the XBB.1.5 spike protein receptor binding domain (RBD) that is recognized by serum antibodies from a donor with hybrid immunity, collected prior to the emergence of the XBB.1.5 variant. T cell assays reveal high frequencies of XBB.1.5 spike-specific CD4 and CD8 T cells amongst donors with hybrid immunity, with the CD4 T cells skewed towards a Th1 cell phenotype and having attenuated effector cytokine secretion as compared to ancestral spike protein-specific cells. Thus, while the XBB.1.5 variant has retained efficient human receptor binding and gained antigenic alterations, it remains susceptible to recognition by T cells induced via vaccination and previous infection. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43326.map.gz emd_43326.map.gz | 50.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43326-v30.xml emd-43326-v30.xml emd-43326.xml emd-43326.xml | 27 KB 27 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_43326.png emd_43326.png | 13.8 KB | ||
| Filedesc metadata |  emd-43326.cif.gz emd-43326.cif.gz | 4.9 KB | ||
| Others |  emd_43326_additional_1.map.gz emd_43326_additional_1.map.gz emd_43326_additional_2.map.gz emd_43326_additional_2.map.gz emd_43326_additional_3.map.gz emd_43326_additional_3.map.gz emd_43326_additional_4.map.gz emd_43326_additional_4.map.gz emd_43326_additional_5.map.gz emd_43326_additional_5.map.gz emd_43326_half_map_1.map.gz emd_43326_half_map_1.map.gz emd_43326_half_map_2.map.gz emd_43326_half_map_2.map.gz | 4.1 MB 4 MB 4.1 MB 4.1 MB 4.1 MB 95.6 MB 95.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43326 http://ftp.pdbj.org/pub/emdb/structures/EMD-43326 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43326 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43326 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_43326.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43326.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Negative Stain EM reconstruction of XBB.1.5 spike protein when mixed with polyclonal fabs from donor 4 (apo - C1 symmetry) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.9 Å | ||||||||||||||||||||||||||||||||||||
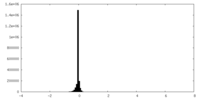
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Negative Stain EM reconstruction of WT spike protein...
| File | emd_43326_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Negative Stain EM reconstruction of WT spike protein when mixed with polyclonal fabs from donor 4 (RBD up directed - C1 symmetry) | ||||||||||||
| Projections & Slices |
| ||||||||||||
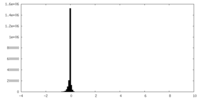
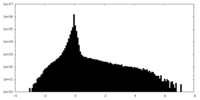
| Density Histograms |
-Additional map: Negative Stain EM reconstruction of XBB.1.5 spike protein...
| File | emd_43326_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Negative Stain EM reconstruction of XBB.1.5 spike protein when mixed with polyclonal IgG from donor 4 (RBD up directed - C1 symmetry) | ||||||||||||
| Projections & Slices |
| ||||||||||||
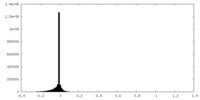
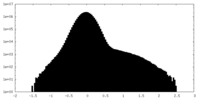
| Density Histograms |
-Additional map: Negative Stain EM reconstruction of WT spike protein...
| File | emd_43326_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Negative Stain EM reconstruction of WT spike protein when mixed with polyclonal fabs from donor 4 (RBD up directed - C1 symmetry) | ||||||||||||
| Projections & Slices |
| ||||||||||||
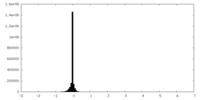
| Density Histograms |
-Additional map: Negative Stain EM reconstruction of WT spike protein...
| File | emd_43326_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Negative Stain EM reconstruction of WT spike protein when mixed with polyclonal fabs from donor 4 (RBD up directed x 2 - C1 symmetry) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Negative Stain EM reconstruction of WT spike protein...
| File | emd_43326_additional_5.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Negative Stain EM reconstruction of WT spike protein when mixed with polyclonal fabs from donor 4 (NTD bottom face directed - C1 symmetry) | ||||||||||||
| Projections & Slices |
| ||||||||||||
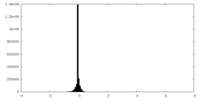
| Density Histograms |
-Half map: #2
| File | emd_43326_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_43326_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 XBB.1.5 spike protein trimer
| Entire | Name: SARS-CoV-2 XBB.1.5 spike protein trimer |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 XBB.1.5 spike protein trimer
| Supramolecule | Name: SARS-CoV-2 XBB.1.5 spike protein trimer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 20.0 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 82636 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)