+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human Complement C3b in complex with Trypanosoma brucei ISG65. | |||||||||
 Map data Map data | Human Complement C3 in complex with T. brucei ISG65. Composite map from local alignments of the CUB-TED-ISG65 and remaining C3b regions. Postprocessed in DeepEMhancer. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complement system / parasite virulence / trypanosome surface protein / host-pathogen complex / IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology informationC5L2 anaphylatoxin chemotactic receptor binding / oviduct epithelium development / regulation of triglyceride biosynthetic process / positive regulation of activation of membrane attack complex / vertebrate eye-specific patterning / positive regulation of apoptotic cell clearance / complement-mediated synapse pruning / Alternative complement activation / positive regulation of phagocytosis, engulfment / Activation of C3 and C5 ...C5L2 anaphylatoxin chemotactic receptor binding / oviduct epithelium development / regulation of triglyceride biosynthetic process / positive regulation of activation of membrane attack complex / vertebrate eye-specific patterning / positive regulation of apoptotic cell clearance / complement-mediated synapse pruning / Alternative complement activation / positive regulation of phagocytosis, engulfment / Activation of C3 and C5 / positive regulation of lipid storage / positive regulation of G protein-coupled receptor signaling pathway / positive regulation of type IIa hypersensitivity / complement-dependent cytotoxicity / complement receptor mediated signaling pathway / positive regulation of D-glucose transmembrane transport / complement activation, alternative pathway / complement activation / endopeptidase inhibitor activity / neuron remodeling / amyloid-beta clearance / B cell activation / positive regulation of vascular endothelial growth factor production / complement activation, classical pathway / Purinergic signaling in leishmaniasis infection / Regulation of Complement cascade / Peptide ligand-binding receptors / Post-translational protein phosphorylation / response to bacterium / fatty acid metabolic process / positive regulation of receptor-mediated endocytosis / Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) / positive regulation of protein phosphorylation / Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell / positive regulation of angiogenesis / azurophil granule lumen / secretory granule lumen / blood microparticle / G alpha (i) signalling events / immune response / G protein-coupled receptor signaling pathway / endoplasmic reticulum lumen / inflammatory response / receptor ligand activity / signaling receptor binding / Neutrophil degranulation / cell surface / signal transduction / protein-containing complex / extracellular space / extracellular exosome / extracellular region / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Cook AD / Higgins MK | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2024 Journal: Elife / Year: 2024Title: Molecular mechanism of complement inhibition by the trypanosome receptor ISG65. Authors: Alexander D Cook / Mark Carrington / Matthew K Higgins /  Abstract: African trypanosomes replicate within infected mammals where they are exposed to the complement system. This system centres around complement C3, which is present in a soluble form in serum but ...African trypanosomes replicate within infected mammals where they are exposed to the complement system. This system centres around complement C3, which is present in a soluble form in serum but becomes covalently deposited onto the surfaces of pathogens after proteolytic cleavage to C3b. Membrane-associated C3b triggers different complement-mediated effectors which promote pathogen clearance. To counter complement-mediated clearance, African trypanosomes have a cell surface receptor, ISG65, which binds to C3b and which decreases the rate of trypanosome clearance in an infection model. However, the mechanism by which ISG65 reduces C3b function has not been determined. We reveal through cryogenic electron microscopy that ISG65 has two distinct binding sites for C3b, only one of which is available in C3 and C3d. We show that ISG65 does not block the formation of C3b or the function of the C3 convertase which catalyses the surface deposition of C3b. However, we show that ISG65 forms a specific conjugate with C3b, perhaps acting as a decoy. ISG65 also occludes the binding sites for complement receptors 2 and 3, which may disrupt recruitment of immune cells, including B cells, phagocytes, and granulocytes. This suggests that ISG65 protects trypanosomes by combining multiple approaches to dampen the complement cascade. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17209.map.gz emd_17209.map.gz | 118.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17209-v30.xml emd-17209-v30.xml emd-17209.xml emd-17209.xml | 19.2 KB 19.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_17209.png emd_17209.png | 158.7 KB | ||
| Filedesc metadata |  emd-17209.cif.gz emd-17209.cif.gz | 7.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17209 http://ftp.pdbj.org/pub/emdb/structures/EMD-17209 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17209 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17209 | HTTPS FTP |
-Related structure data
| Related structure data |  8ovbMC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17209.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17209.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human Complement C3 in complex with T. brucei ISG65. Composite map from local alignments of the CUB-TED-ISG65 and remaining C3b regions. Postprocessed in DeepEMhancer. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||||||||||||||||||
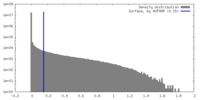
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Complex of Human Complement C3b and T. brucei ISG65
| Entire | Name: Complex of Human Complement C3b and T. brucei ISG65 |
|---|---|
| Components |
|
-Supramolecule #1: Complex of Human Complement C3b and T. brucei ISG65
| Supramolecule | Name: Complex of Human Complement C3b and T. brucei ISG65 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Molecular weight | Theoretical: 44.2 KDa |
-Supramolecule #2: Human Complement C3b
| Supramolecule | Name: Human Complement C3b / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 Details: C3 purified from human serum, C3b generated from C3 by limited trypsin proteolysis. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Tissue: Blood Homo sapiens (human) / Tissue: Blood |
-Supramolecule #3: Trypanosoma brucei ISG65
| Supramolecule | Name: Trypanosoma brucei ISG65 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Complement C3f fragment
| Macromolecule | Name: Complement C3f fragment / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Tissue: Blood Homo sapiens (human) / Tissue: Blood |
| Molecular weight | Theoretical: 71.154047 KDa |
| Sequence | String: SPMYSIITPN ILRLESEETM VLEAHDAQGD VPVTVTVHDF PGKKLVLSSE KTVLTPATNH MGNVTFTIPA NREFKSEKGR NKFVTVQAT FGTQVVEKVV LVSLQSGYLF IQTDKTIYTP GSTVLYRIFT VNHKLLPVGR TVMVNIENPE GIPVKQDSLS S QNQLGVLP ...String: SPMYSIITPN ILRLESEETM VLEAHDAQGD VPVTVTVHDF PGKKLVLSSE KTVLTPATNH MGNVTFTIPA NREFKSEKGR NKFVTVQAT FGTQVVEKVV LVSLQSGYLF IQTDKTIYTP GSTVLYRIFT VNHKLLPVGR TVMVNIENPE GIPVKQDSLS S QNQLGVLP LSWDIPELVN MGQWKIRAYY ENSPQQVFST EFEVKEYVLP SFEVIVEPTE KFYYIYNEKG LEVTITARFL YG KKVEGTA FVIFGIQDGE QRISLPESLK RIPIEDGSGE VVLSRKVLLD GVQNPRAEDL VGKSLYVSAT VILHSGSDMV QAE RSGIPI VTSPYQIHFT KTPKYFKPGM PFDLMVFVTN PDGSPAYRVP VAVQGEDTVQ SLTQGDGVAK LSINTHPSQK PLSI TVRTK KQELSEAEQA TRTMQALPYS TVGNSNNYLH LSVLRTELRP GETLNVNFLL RMDRAHEAKI RYYTYLIMNK GRLLK AGRQ VREPGQDLVV LPLSITTDFI PSFRLVAYYT LIGASGQREV VADSVWVDVK DSCVGSLVVK SGQSEDRQPV PGQQMT LKI EGDHGARVVL VAVDKGVFVL NKKNKLTQSK IWDVVEKADI GCTPGSGKDY AGVFSDAGLT FTSSSGQQTA QRAELQC PQ UniProtKB: Complement C3 |
-Macromolecule #2: Complement C3
| Macromolecule | Name: Complement C3 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Tissue: Blood Homo sapiens (human) / Tissue: Blood |
| Molecular weight | Theoretical: 104.073164 KDa |
| Sequence | String: SNLDEDIIAE ENIVSRSEFP ESWLWNVEDL KEPPKNGIST KLMNIFLKDS ITTWEILAVS MSDKKGICVA DPFEVTVMQD FFIDLRLPY SVVRNEQVEI RAVLYNYRQN QELKVRVELL HNPAFCSLAT TKRRHQQTVT IPPKSSLSVP YVIVPLKTGL Q EVEVKAAV ...String: SNLDEDIIAE ENIVSRSEFP ESWLWNVEDL KEPPKNGIST KLMNIFLKDS ITTWEILAVS MSDKKGICVA DPFEVTVMQD FFIDLRLPY SVVRNEQVEI RAVLYNYRQN QELKVRVELL HNPAFCSLAT TKRRHQQTVT IPPKSSLSVP YVIVPLKTGL Q EVEVKAAV YHHFISDGVR KSLKVVPEGI RMNKTVAVRT LDPERLGREG VQKEDIPPAD LSDQVPDTES ETRILLQGTP VA QMTEDAV DAERLKHLIV TPSGCGEQNM IGMTPTVIAV HYLDETEQWE KFGLEKRQGA LELIKKGYTQ QLAFRQPSSA FAA FVKRAP STWLTAYVVK VFSLAVNLIA IDSQVLCGAV KWLILEKQKP DGVFQEDAPV IHQEMIGGLR NNNEKDMALT AFVL ISLQE AKDICEEQVN SLPGSITKAG DFLEANYMNL QRSYTVAIAG YALAQMGRLK GPLLNKFLTT AKDKNRWEDP GKQLY NVEA TSYALLALLQ LKDFDFVPPV VRWLNEQRYY GGGYGSTQAT FMVFQALAQY QKDAPDHQEL NLDVSLQLPS RSSKIT HRI HWESASLLRS EETKENEGFT VTAEGKGQGT LSVVTMYHAK AKDQLTCNKF DLKVTIKPAP ETEKRPQDAK NTMILEI CT RYRGDQDATM SILDISMMTG FAPDTDDLKQ LANGVDRYIS KYELDKAFSD RNTLIIYLDK VSHSEDDCLA FKVHQYFN V ELIQPGAVKV YAYYNLEESC TRFYHPEKED GKLNKLCRDE LCRCAEENCF IQKSDDKVTL EERLDKACEP GVDYVYKTR LVKVQLSNDF DEYIMAIEQT IKSGSDEVQV GQQRTFISPI KCREALKLEE KKHYLMWGLS SDFWGEKPNL SYIIGKDTWV EHWPEEDEC QDEENQKQCQ DLGAFTESMV VFGCPN UniProtKB: Complement C3 |
-Macromolecule #3: ISG65 G
| Macromolecule | Name: ISG65 G / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 32.597322 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DNRVPGDKNL TKEGAAALCK MKHLADKVAE KRSQELKDRT QNFAGYIEFE LYRIDYWLEK LNGPKGRKDG YAKLSDSDIE KVKEIFDKA KDGIAKQLPE AKKAGEDAEK LHTEVKEAAA NARGQDLDDH KQKSTGLYRV LNWYCITKEE SHNATPNCDG I QFRNHYLS ...String: DNRVPGDKNL TKEGAAALCK MKHLADKVAE KRSQELKDRT QNFAGYIEFE LYRIDYWLEK LNGPKGRKDG YAKLSDSDIE KVKEIFDKA KDGIAKQLPE AKKAGEDAEK LHTEVKEAAA NARGQDLDDH KQKSTGLYRV LNWYCITKEE SHNATPNCDG I QFRNHYLS VNRSAIDCSS TGYEENYDWS ANALQVALNS WENVKPKKLE SAGSDENCNI GQSSESHPCT MTEEWQTHYK ET VKKLKEL EGAHEKGRRA HDAMLGYANT AYAVNTKVEQ EKPLAEVIAA AK UniProtKB: ISG65 G |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.2 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 278.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 14339 / Average exposure time: 3.03 sec. / Average electron dose: 49.9 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)