7Z06
 
 | |
8CRL
 
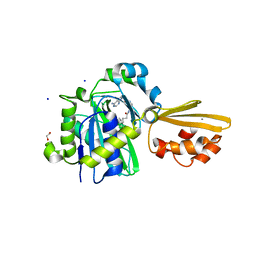 | | Crystal structure of LplA1 in complex with the inhibitor C3 (Listeria monocytogenes) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, SODIUM ION, ... | | Authors: | Dienemann, J.-N, Chen, S.-Y, Hitzenberger, M, Sievert, M.L, Hacker, S.M, Prigge, S.T, Zacharias, M, Groll, M, Sieber, S.A. | | Deposit date: | 2023-03-08 | | Release date: | 2023-06-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Chemical Proteomic Strategy Reveals Inhibitors of Lipoate Salvage in Bacteria and Parasites.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8CRI
 
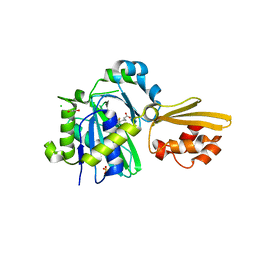 | | Crystal structure of LplA1 in complex with lipoic acid (Listeria monocytogenes) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, LIPOIC ACID, ... | | Authors: | Dienemann, J.-N, Chen, S.-Y, Hitzenberger, M, Sievert, M.L, Hacker, S.M, Prigge, S.T, Zacharias, M, Groll, M, Sieber, S.A. | | Deposit date: | 2023-03-08 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Chemical Proteomic Strategy Reveals Inhibitors of Lipoate Salvage in Bacteria and Parasites.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8CRJ
 
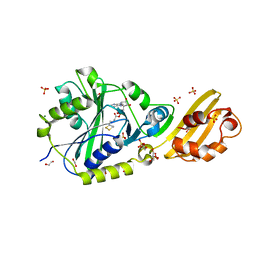 | | Crystal structure of LplA1 in complex with lipoyl-AMP (Listeria monocytogenes) | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[(R)-({5-[(3R)-1,2-DITHIOLAN-3-YL]PENTANOYL}OXY)(HYDROXY)PHOSPHORYL]ADENOSINE, GLYCEROL, ... | | Authors: | Dienemann, J.-N, Chen, S.-Y, Hitzenberger, M, Sievert, M.L, Hacker, S.M, Prigge, S.T, Zacharias, M, Groll, M, Sieber, S.A. | | Deposit date: | 2023-03-08 | | Release date: | 2023-06-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Chemical Proteomic Strategy Reveals Inhibitors of Lipoate Salvage in Bacteria and Parasites.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8OLL
 
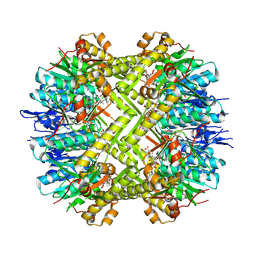 | | Staphylococcus aureus ClpP in complex with the natural product beta-lactone inhibitor Cystargolide A at 2.7 A resolution | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-03-30 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8OLR
 
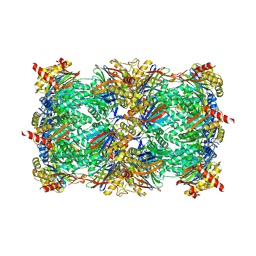 | | Structure of yeast 20S proteasome in complex with the natural product beta-lactone inhibitor Cystargolide A | | Descriptor: | CHLORIDE ION, Cystargolide A (bound), MAGNESIUM ION, ... | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-03-30 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8R03
 
 | | Staphylococcus aureus ClpP in complex with the natural product beta-lactone inhibitor Cystargolide A at 2.0 A resolution | | Descriptor: | 1,2-ETHANEDIOL, ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Liebhart, E, Kuttenlochner, W, Kulik, A, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-10-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8R05
 
 | | Photorhabdus lamondii ClpP in complex with the natural product beta-lactone inhibitor Cystargolide A at 2.5 A resolution | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Liebhart, E, Kuttenlochner, W, Kulik, A, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-10-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8R04
 
 | | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active Site Inhibitor Cystargolide A | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Liebhart, E, Kuttenlochner, W, Kulik, A, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-10-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
3QWD
 
 | | Crystal structure of ClpP from Staphylococcus aureus | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, CHLORIDE ION | | Authors: | Geiger, S.R, Boettcher, T, Sieber, S.A, Cramer, P. | | Deposit date: | 2011-02-28 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Conformational Switch Underlies ClpP Protease Function.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
4LQI
 
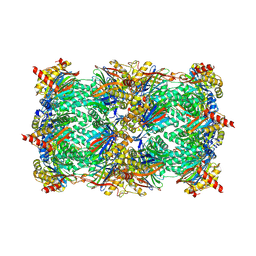 | | Yeast 20S Proteasome in complex with Vibralactone | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | List, A, Zeiler, E, Gallastegui, N, Rusch, M, Hedberg, C, Sieber, S.A, Groll, M. | | Deposit date: | 2013-07-18 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Omuralide and Vibralactone: Differences in the Proteasome-beta-Lactone-gamma-Lactam Binding Scaffold Alter Target Preferences.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4MXI
 
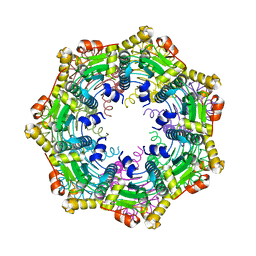 | | ClpP Ser98dhA | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | Gersch, M, Kolb, R, Alte, F, Groll, M, Sieber, S.A. | | Deposit date: | 2013-09-26 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Disruption of Oligomerization and Dehydroalanine Formation as Mechanisms for ClpP Protease Inhibition.
J.Am.Chem.Soc., 136, 2014
|
|
4R18
 
 | | Ligand-induced Lys33-Thr1 crosslinking at subunit beta5 of the yeast 20S proteasome | | Descriptor: | ALPHA-AMINOBUTYRIC ACID, MAGNESIUM ION, PROTEASOME SUBUNIT ALPHA TYPE-1, ... | | Authors: | Dubiella, C, Cui, H, Gersch, M, Brouwer, A.J, Sieber, S.A, Krueger, A, Liskamp, R, Groll, M. | | Deposit date: | 2014-08-04 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Selective inhibition of the immunoproteasome by ligand-induced crosslinking of the active site.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4RYF
 
 | | ClpP1/2 heterocomplex from Listeria monocytogenes | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, MALONATE ION, SODIUM ION | | Authors: | Dahmen, M, Vielberg, M.-T, Groll, M, Sieber, S.A. | | Deposit date: | 2014-12-15 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and mechanism of the caseinolytic protease ClpP1/2 heterocomplex from Listeria monocytogenes.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4R17
 
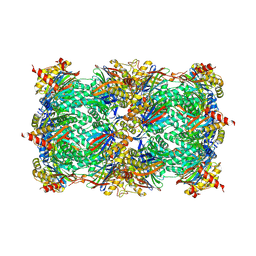 | | Ligand-induced aziridine-formation at subunit beta5 of the yeast 20S proteasome | | Descriptor: | (2S,3S)-3-methylaziridine-2-carboxylic acid, MAGNESIUM ION, Proteasome subunit alpha type-1, ... | | Authors: | Dubiella, C, Cui, H, Gersch, M, Brouwer, A.J, Sieber, S.A, Krueger, A, Liskamp, R, Groll, M. | | Deposit date: | 2014-08-04 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Selective inhibition of the immunoproteasome by ligand-induced crosslinking of the active site.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
2M0G
 
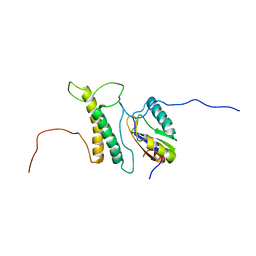 | | Structure, phosphorylation and U2AF65 binding of the Nterminal Domain of splicing factor 1 during 3 splice site Recognition | | Descriptor: | Splicing factor 1, Splicing factor U2AF 65 kDa subunit | | Authors: | Madl, T, Sattler, M, Zhang, Y, Bagdiul, I, Kern, T, Kang, H, Zou, P, Maeusbacher, N, Sieber, S.A, Kraemer, A. | | Deposit date: | 2012-10-25 | | Release date: | 2013-01-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure, phosphorylation and U2AF65 binding of the N-terminal domain of splicing factor 1 during 3'-splice site recognition.
Nucleic Acids Res., 41, 2013
|
|
2M09
 
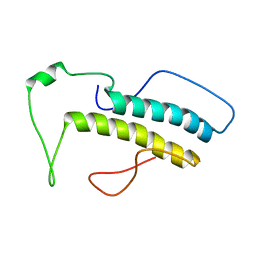 | | Structure, phosphorylation and U2AF65 binding of the Nterminal Domain of splicing factor 1 during 3 splice site Recognition | | Descriptor: | Splicing factor 1 | | Authors: | Madl, T, Sattler, M, Zhang, Y, Bagdiul, I, Kern, T, Kang, H, Zou, P, Maeusbacher, N, Sieber, S.A, Kraemer, A. | | Deposit date: | 2012-10-22 | | Release date: | 2013-01-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, phosphorylation and U2AF65 binding of the N-terminal domain of splicing factor 1 during 3'-splice site recognition.
Nucleic Acids Res., 41, 2013
|
|
4C5L
 
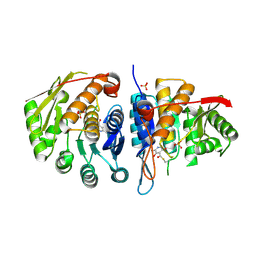 | | Structure of the pyridoxal kinase from Staphylococcus aureus in complex with pyridoxal | | Descriptor: | 3-HYDROXY-5-(HYDROXYMETHYL)-2-METHYLISONICOTINALDEHYDE, 4,5-bis(hydroxymethyl)-2-methyl-pyridin-3-ol, PHOSPHOMETHYLPYRIMIDINE KINASE, ... | | Authors: | Nodwell, M, Alte, F, Sieber, S.A, Schneider, S. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Subfamily of Bacterial Ribokinases Utilizes a Hemithioacetal for Pyridoxal Phosphate Salvage.
J.Am.Chem.Soc., 136, 2014
|
|
4C5N
 
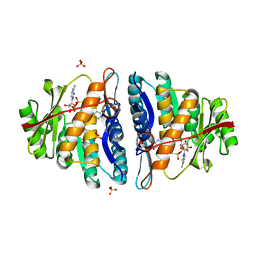 | | Structure of the pyridoxal kinase from Staphylococcus aureus in complex with AMP-PCP and pyridoxal | | Descriptor: | 3-HYDROXY-5-(HYDROXYMETHYL)-2-METHYLISONICOTINALDEHYDE, 4,5-bis(hydroxymethyl)-2-methyl-pyridin-3-ol, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Nodwell, M, Alte, F, Sieber, S.A, Schneider, S. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Subfamily of Bacterial Ribokinases Utilizes a Hemithioacetal for Pyridoxal Phosphate Salvage.
J.Am.Chem.Soc., 136, 2014
|
|
4C5M
 
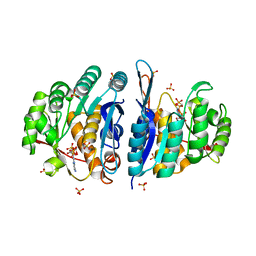 | | Structure of the pyridoxal kinase from Staphylococcus aureus in complex with AMP-PCP | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, PHOSPHOMETHYLPYRIMIDINE KINASE, SULFATE ION | | Authors: | Nodwell, M, Alte, F, Sieber, S.A, Schneider, S. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A Subfamily of Bacterial Ribokinases Utilizes a Hemithioacetal for Pyridoxal Phosphate Salvage.
J.Am.Chem.Soc., 136, 2014
|
|
4C5K
 
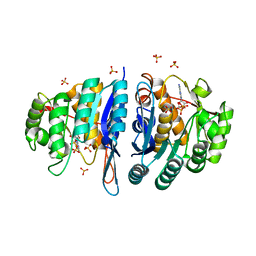 | | Structure of the pyridoxal kinase from Staphylococcus aureus in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PHOSPHOMETHYLPYRIMIDINE KINASE, SULFATE ION | | Authors: | Nodwell, M, Alte, F, Sieber, S.A, Schneider, S. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Subfamily of Bacterial Ribokinases Utilizes a Hemithioacetal for Pyridoxal Phosphate Salvage.
J.Am.Chem.Soc., 136, 2014
|
|
4C5J
 
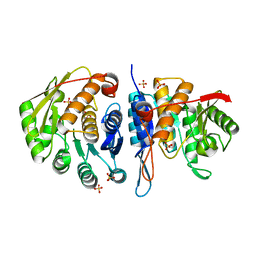 | | Structure of the pyridoxal kinase from Staphylococcus aureus | | Descriptor: | PHOSPHOMETHYLPYRIMIDINE KINASE, SULFATE ION | | Authors: | Nodwell, M, Alte, F, Sieber, S.A, Schneider, S. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A Subfamily of Bacterial Ribokinases Utilizes a Hemithioacetal for Pyridoxal Phosphate Salvage.
J.Am.Chem.Soc., 136, 2014
|
|
5AC2
 
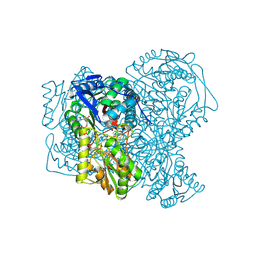 | | human aldehyde dehydrogenase 1A1 with duocarmycin analog | | Descriptor: | 1-[(1S)-1-methyl-5-oxidanyl-1,2-dihydrobenzo[e]indol-3-yl]hexan-1-one, RETINAL DEHYDROGENASE 1, YTTERBIUM (III) ION, ... | | Authors: | Koch, M.F, Harteis, S, Blank, I.D, Pestel, G, Tietze, L.F, Ochsenfeld, C, Schneider, S, Sieber, S.A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural, Biochemical, and Computational Studies Reveal the Mechanism of Selective Aldehyde Dehydrogenase 1A1 Inhibition by Cytotoxic Duocarmycin Analogues.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5AC0
 
 | | ovis aries Aldehyde Dehydrogenase 1A1 in complex with a duocarmycin analog | | Descriptor: | 1-[(1S)-1-methyl-5-oxidanyl-1,2-dihydrobenzo[e]indol-3-yl]hexan-1-one, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Koch, M.F, Harteis, S, Blank, I.D, Pestel, G, Tietze, L.F, Ochsenfeld, C, Schneider, S, Sieber, S.A. | | Deposit date: | 2015-08-10 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural, Biochemical, and Computational Studies Reveal the Mechanism of Selective Aldehyde Dehydrogenase 1A1 Inhibition by Cytotoxic Duocarmycin Analogues.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5AC1
 
 | | Sheep aldehyde dehydrogenase 1A1 with duocarmycin analog inhibitor | | Descriptor: | 1-[(1S)-1-methyl-5-oxidanyl-1,2-dihydrobenzo[e]indol-3-yl]hexan-1-one, MAGNESIUM ION, RETINAL DEHYDROGENASE 1, ... | | Authors: | Koch, M.F, Harteis, S, Blank, I.D, Pestel, G, Tietze, L.F, Ochsenfeld, C, Schneider, S, Sieber, S.A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural, Biochemical, and Computational Studies Reveal the Mechanism of Selective Aldehyde Dehydrogenase 1A1 Inhibition by Cytotoxic Duocarmycin Analogues.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
