4DRT
 
 | | Three dimensional structure of de novo designed serine hydrolase OSH26, Northeast Structural Genomics Consortium (NESG) target OR89 | | Descriptor: | CHLORIDE ION, SODIUM ION, de novo designed serine hydrolase, ... | | Authors: | Kuzin, A, Su, M, Rajagopalan, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-02-17 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4ESS
 
 | | Crystal Structure of E6D/L155R variant of de novo designed serine hydrolase OSH55, Northeast Structural Genomics Consortium (NESG) Target OR187 | | Descriptor: | OR187 | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Kornhaber, K, Kornhaber, G, Rajagopalan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-23 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9971 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4ETJ
 
 | | Crystal Structure of E6H variant of de novo designed serine hydrolase OSH55, Northeast Structural Genomics Consortium (NESG) Target OR185 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, CHLORIDE ION, ... | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Kornhaber, K, Kornhaber, G, Rajagopalan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
3TP4
 
 | | Crystal Structure of engineered protein at the resolution 1.98A, Northeast Structural Genomics Consortium Target OR128 | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Rajagopalan, S, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-07 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
3V45
 
 | | Crystal Structure of de novo designed serine hydrolase OSH55, Northeast Structural Genomics Consortium Target OR130 | | Descriptor: | CHLORIDE ION, SODIUM ION, Serine hydrolase OSH55 | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Maglaqui, M, Xiao, R, Kohan, E, Rajagopalan, S, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-12-14 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
3T03
 
 | | Crystal structure of PPAR gamma ligand binding domain in complex with a novel partial agonist GQ-16 | | Descriptor: | (5Z)-5-(5-bromo-2-methoxybenzylidene)-3-(4-methylbenzyl)-1,3-thiazolidine-2,4-dione, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Rajagopalan, S, Webb, P, Baxter, J.D, Brennan, R.G, Phillips, K.J. | | Deposit date: | 2011-07-19 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | GQ-16, a novel peroxisome proliferator-activated receptor (PPAR gamma) ligand, promotes insulin sensitization without weight gain.
J.Biol.Chem., 287, 2012
|
|
4JVV
 
 | | Crystal structure of the evolved variant of the computationally designed serine hydrolase, OSH55.4_H1, covalently bound with diisopropyl fluorophosphate (DFP), Northeast Structural Genomics Consortium (NESG) Target OR273 | | Descriptor: | evolved variant of the computationally designed serine hydrolase | | Authors: | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Tong, S, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-03-26 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.288 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4JCA
 
 | | Crystal Structure of the apo form of the evolved variant of the computationally designed serine hydrolase, OSH55.4_H1. Northeast Structural Genomics Consortium (NESG) Target OR273 | | Descriptor: | CITRIC ACID, RUBIDIUM ION, serine hydrolase | | Authors: | Kuzin, A.P, Lew, S, Rajagopalan, S, Seetharaman, J, Tong, S, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-02-21 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.411 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4JLL
 
 | | Crystal Structure of the evolved variant of the computationally designed serine hydrolase, OSH55.4_H1 covalently bound with FP-alkyne, Northeast Structural Genomics Consortium (NESG) Target OR273 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Tong, S, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-03-12 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4F2V
 
 | | Crystal Structure of de novo designed serine hydrolase, Northeast Structural Genomics Consortium (NESG) Target OR165 | | Descriptor: | DI(HYDROXYETHYL)ETHER, DODECYL-ALPHA-D-MALTOSIDE, De novo designed serine hydrolase | | Authors: | Kuzin, A, Lew, S, Seetharaman, J, Maglaqui, M, Xiao, R, Kohan, E, Rajagopalan, S, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-05-08 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4ETK
 
 | | Crystal Structure of E6A/L130D/A155H variant of de novo designed serine hydrolase, Northeast Structural Genomics Consortium (NESG) Target OR186 | | Descriptor: | De novo designed serine hydrolase, SODIUM ION | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Kornhaber, K, Kornhaber, G, Rajagopalan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4KYB
 
 | | Crystal Structure of de novo designed serine hydrolase OSH55.14_E3, Northeast Structural Genomics Consortium Target OR342 | | Descriptor: | Designed Protein OR342, PHOSPHATE ION | | Authors: | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Mao, L, Xiao, R, Lee, D, Raja, S, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-28 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.909 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR342
To be Published
|
|
4N3P
 
 | | Crystal Structure of De Novo designed Serine Hydrolase OSH18, Northeast Structural Genomics Consortium (NESG) Target OR396 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Mao, L, Xiao, R, Kogan, S, Maglaqui, M, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-10-07 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Crystal Structure of De Novo designed Serine Hydrolase OSH18, Northeast Structural Genomics Consortium (NESG) Target OR396
To be Published
|
|
4GVV
 
 | | Crystal Structure of de novo design serine hydrolase OSH55.27, Northeast Structural Genomics Consortium (NESG) Target OR246 | | Descriptor: | De novo design serine hydrolase | | Authors: | Kuzin, A, Lew, S, Seetharaman, J, Mao, M, Xiao, R, Kohan, E, Rajagopalan, S, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-08-31 | | Release date: | 2012-09-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.895 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR246
To be Published
|
|
4GVW
 
 | | Three-dimensional structure of the de novo designed serine hydrolase 2bfq_3, Northeast Structural Genomics Consortium (NESG) Target OR248 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETIC ACID, De novo designed serine hydrolase, ... | | Authors: | Kuzin, A, Lew, S, Seetharaman, J, Rajagopalan, S, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-08-31 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.113 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR248
To be Published
|
|
4JGK
 
 | | Crystal Structure of the evolved variant of the computationally designed serine hydrolase, Northeast Structural Genomics Consortium (NESG) Target OR275 | | Descriptor: | evolved variant of a designed serine hydrolase | | Authors: | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Mao, L, Xiao, R, Lee, D, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-03-01 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.883 Å) | | Cite: | Crystal Structure of the evolved variant of the computationally designed serine hydrolase, Northeast Structural Genomics Consortium (NESG) Target OR275
To be Published
|
|
4JBC
 
 | | Crystal Structure of the computationally designed serine hydrolase 3mmj_2, Northeast Structural Genomics Consortium (NESG) Target OR318 | | Descriptor: | PHOSPHATE ION, designed serine hydrolase 3mmj_2 | | Authors: | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Maglaqui, M, Xiao, R, Lee, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Baker, D, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-02-19 | | Release date: | 2013-03-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Crystal Structure of the computationally designed serine hydrolase 3mmj_2, Northeast Structural Genomics Consortium (NESG) Target OR318
To be Published
|
|
4J4Z
 
 | | Crystal structure of the improved variant of the evolved serine hydrolase, OSH55.4_H1.2, bond with sulfate ion in the active site, Northeast Structural Genomics Consortium (NESG) Target OR301 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, Designed serine hydrolase variant OSH55.4_H1.2, ... | | Authors: | Kuzin, A.P, Lew, S, Rajagopalan, S, Maglaqui, M, Xiao, R, Lee, D, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-02-07 | | Release date: | 2013-03-06 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal structure of the improved variant of the evolved serine hydrolase, OSH55.4_H1.2, bond with sulfate ion in the active site, Northeast Structural Genomics Consortium (NESG) Target OR301
To be Published
|
|
4K0C
 
 | | Crystal Structure of the computationally designed serine hydrolase. Northeast Structural Genomics Consortium (NESG) Target OR317 | | Descriptor: | designed serine hydrolase | | Authors: | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Maglaqui, M, Xiao, R, Lee, D, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-04-03 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR317
To be Published
|
|
4HF2
 
 | |
4HF0
 
 | | Crystal Structure of Apo IscR | | Descriptor: | HTH-type transcriptional regulator IscR, SULFATE ION | | Authors: | Rajagopalan, S.R, Phillips, K.J. | | Deposit date: | 2012-10-04 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Studies of IscR reveal a unique mechanism for metal-dependent regulation of DNA binding specificity.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4HF1
 
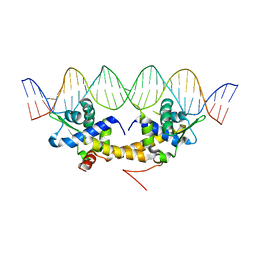 | | Crystal Structure of IscR bound to its promoter | | Descriptor: | DNA (29-MER), HTH-type transcriptional regulator IscR | | Authors: | Rajagopalan, S.R, Phillips, K.J. | | Deposit date: | 2012-10-04 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.222 Å) | | Cite: | Studies of IscR reveal a unique mechanism for metal-dependent regulation of DNA binding specificity.
Nat.Struct.Mol.Biol., 20, 2013
|
|
6CHW
 
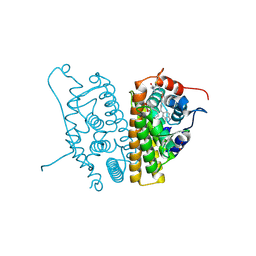 | | Estrogen Receptor Alpha Y537S covalently bound to antagonist H3B-5942. | | Descriptor: | 1,2-ETHANEDIOL, 4-[(2-{4-[(1E)-1-(1H-indazol-5-yl)-2-phenylbut-1-en-1-yl]phenoxy}ethyl)amino]-N,N-dimethylbutanamide, DIMETHYL SULFOXIDE, ... | | Authors: | Larsen, N.A. | | Deposit date: | 2018-02-23 | | Release date: | 2018-03-21 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Discovery of Selective Estrogen Receptor Covalent Antagonists for the Treatment of ER alphaWTand ER alphaMUTBreast Cancer.
Cancer Discov, 8, 2018
|
|
6CHZ
 
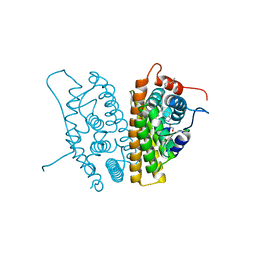 | | Estrogen Receptor Alpha Y537S bound to antagonist H3B-9224. | | Descriptor: | 1,2-ETHANEDIOL, 4-[(2-{4-[(1E)-1-(1H-indazol-5-yl)-2-phenylbut-1-en-1-yl]phenoxy}ethyl)amino]-N,N-dimethylbutanamide, Estrogen receptor | | Authors: | Larsen, N.A. | | Deposit date: | 2018-02-23 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Discovery of Selective Estrogen Receptor Covalent Antagonists for the Treatment of ER alphaWTand ER alphaMUTBreast Cancer.
Cancer Discov, 8, 2018
|
|
4QL8
 
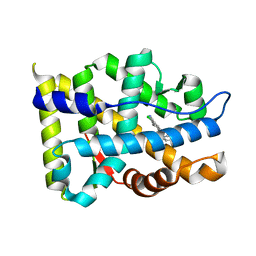 | | Crystal structure of Androgen Receptor in complex with the ligand | | Descriptor: | 2-chloro-4-[(3S,3aS,4S)-4-hydroxy-3-methoxy-3a,4,5,6-tetrahydro-3H-pyrrolo[1,2-b]pyrazol-2-yl]-3-methylbenzonitrile, Androgen receptor | | Authors: | Krishnamurthy, N, Sangeetha, R, Ghadiyaram, C, Sasmal, S, Subramanya, H.S. | | Deposit date: | 2014-06-11 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 3-alkoxy-pyrrolo[1,2-b]pyrazolines as selective androgen receptor modulators with ideal physicochemical properties for transdermal administration
J.Med.Chem., 57, 2014
|
|
