1T3S
 
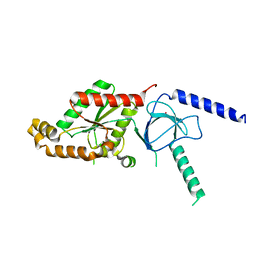 | | Structural Analysis of the Voltage-Dependent Calcium Channel Beta Subunit Functional Core | | Descriptor: | Dihydropyridine-sensitive L-type, calcium channel beta-2 subunit, MERCURY (II) ION | | Authors: | Opatowsky, Y, Chen, C.-C, Campbell, K.P, Hirsch, J.A. | | Deposit date: | 2004-04-27 | | Release date: | 2004-05-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of the voltage-dependent calcium channel beta subunit functional core and its complex with the alpha 1 interaction domain.
Neuron, 42, 2004
|
|
1T3L
 
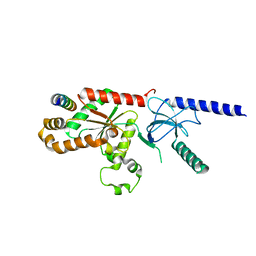 | | Structural Analysis of the Voltage-Dependent Calcium Channel Beta Subunit Functional Core in Complex with Alpha1 Interaction Domain | | Descriptor: | Dihydropyridine-sensitive L-type, calcium channel beta-2 subunit, Voltage-dependent L-type calcium channel alpha-1S subunit | | Authors: | Opatowsky, Y, Chen, C.-C, Campbell, K.P, Hirsch, J.A. | | Deposit date: | 2004-04-27 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analysis of Voltage-Dependent Calcium Channel Beta Subunit Functional Core and Its Complex with the Alpha1 Interaction Domain
NEURON, 42, 2004
|
|
2HT6
 
 | |
4PQJ
 
 | |
9QZK
 
 | | SadB of Pseudomonas aeruginosa | | Descriptor: | HDOD domain-containing protein | | Authors: | Opatowsky, Y, Sporny, M. | | Deposit date: | 2025-04-23 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | SadB, a mediator of AmrZ proteolysis and biofilm development in Pseudomonas aeruginosa.
NPJ Biofilms Microbiomes, 11, 2025
|
|
5NOI
 
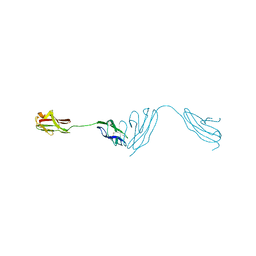 | |
4RTT
 
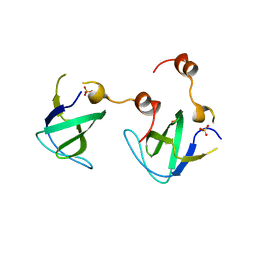 | |
4RUG
 
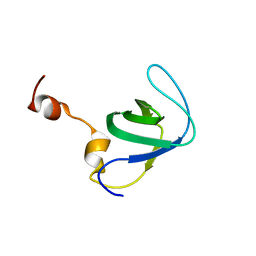 | |
6ZG0
 
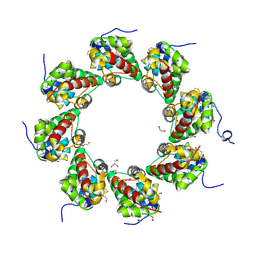 | | SARM1 SAM1-2 domains | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sporny, M, Guez-Haddad, J, Khazma, T, Yaron, A, Dessau, M, Mim, C, Isupov, M.N, Zalk, R, Hons, M, Opatowsky, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2020-11-11 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structural basis for SARM1 inhibition and activation under energetic stress.
Elife, 9, 2020
|
|
3KVQ
 
 | |
8P2M
 
 | | C. elegans TIR-1 protein. | | Descriptor: | NAD(+) hydrolase tir-1 | | Authors: | Isupov, M.N, Opatowsky, Y. | | Deposit date: | 2023-05-16 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structure-function analysis of ceTIR-1/hSARM1 explains the lack of Wallerian axonal degeneration in C. elegans.
Cell Rep, 42, 2023
|
|
8P2L
 
 | |
6QWV
 
 | | SARM1 SAM1-2 domains | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sporny, M, Isupov, N.M, Opatowsky, Y. | | Deposit date: | 2019-03-06 | | Release date: | 2019-07-03 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural Evidence for an Octameric Ring Arrangement of SARM1.
J.Mol.Biol., 431, 2019
|
|
4OMB
 
 | | Phosphate binding protein | | Descriptor: | PHOSPHATE ION, PLATINUM (II) ION, Phosphate binding protein | | Authors: | Neznansky, A, Opatowsky, Y. | | Deposit date: | 2014-01-27 | | Release date: | 2014-12-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Pseudomonas aeruginosa phosphate transport protein PstS plays a phosphate-independent role in biofilm formation.
Faseb J., 28, 2014
|
|
4HLJ
 
 | | Axon Guidance Receptor | | Descriptor: | PENTAETHYLENE GLYCOL, Roundabout homolog 1 | | Authors: | Barak, R, Opatowsky, Y. | | Deposit date: | 2012-10-17 | | Release date: | 2013-11-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Axon Guidance Receptor
To be Published
|
|
6ZFX
 
 | | hSARM1 GraFix-ed | | Descriptor: | (~{E})-4-methylnon-4-enedial, NAD(+) hydrolase SARM1 | | Authors: | Sporny, M, Guez-Haddad, J, Khazma, T, Yaron, A, Dessau, M, Mim, C, Isupov, M.N, Zalk, R, Hons, M, Opatowsky, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural basis for SARM1 inhibition and activation under energetic stress.
Elife, 9, 2020
|
|
6ZG1
 
 | | SARM1 SAM1-2 domains | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sporny, M, Guez-Haddad, J, Khazma, T, Yaron, A, Dessau, M, Mim, C, Isupov, M.N, Zalk, R, Hons, M, Opatowsky, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2020-11-11 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural basis for SARM1 inhibition and activation under energetic stress.
Elife, 9, 2020
|
|
7ANW
 
 | | hSARM1 NAD+ complex | | Descriptor: | NAD(+) hydrolase SARM1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Sporny, M, Guez-Haddad, J, Khazma, T, Yaron, A, Mim, C, Isupov, M.N, Zalk, R, Dessau, M, Hons, M, Opatowsky, Y. | | Deposit date: | 2020-10-13 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural basis for SARM1 inhibition and activation under energetic stress.
Elife, 9, 2020
|
|
6IAA
 
 | | hRobo2 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Roundabout homolog 2, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Barak, R, Isupov, N.M, Opatowsky, Y. | | Deposit date: | 2018-11-26 | | Release date: | 2019-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural Principles in Robo Activation and Auto-inhibition.
Cell, 177, 2019
|
|
6I9S
 
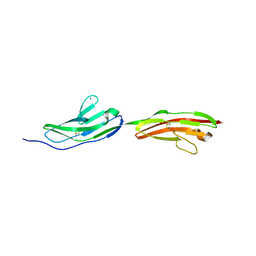 | |
5I6J
 
 | |
5I7D
 
 | |
5I6R
 
 | | Crystal Structure of srGAP2 F-BARx WT Form-1 | | Descriptor: | ACETATE ION, D-MALATE, SLIT-ROBO Rho GTPase-activating protein 2, ... | | Authors: | Sporny, M, Guez-Haddad, J, Isupov, M.N, Opatowsky, Y. | | Deposit date: | 2016-02-16 | | Release date: | 2017-08-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis for srGAP2 Membrane Interactions, and Antagonism by the Human Specific Paralog srGAP2C
To Be Published
|
|
2EC8
 
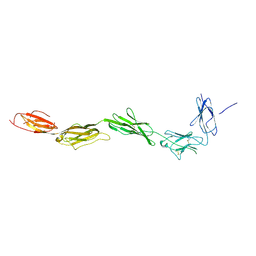 | | Crystal structure of the exctracellular domain of the receptor tyrosine kinase, Kit | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Mast/stem cell growth factor receptor | | Authors: | Yuzawa, S, Opatowsky, Y, Zhang, Z, Mandiyan, V, Lax, I, Schlessinger, J. | | Deposit date: | 2007-02-11 | | Release date: | 2007-08-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Activation of the Receptor Tyrosine Kinase KIT by Stem Cell Factor
Cell(Cambridge,Mass.), 130, 2007
|
|
2E9W
 
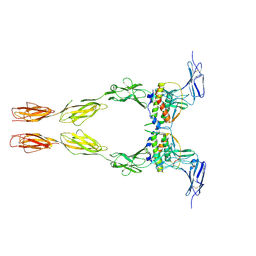 | | Crystal structure of the extracellular domain of Kit in complex with stem cell factor (SCF) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Kit ligand, Mast/stem cell growth factor receptor | | Authors: | Yuzawa, S, Opatowsky, Y, Zhang, Z, Mandiyan, V, Lax, I, Schlessinger, J. | | Deposit date: | 2007-01-27 | | Release date: | 2007-08-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Basis for Activation of the Receptor Tyrosine Kinase KIT by Stem Cell Factor
Cell(Cambridge,Mass.), 130, 2007
|
|
