1ZFD
 
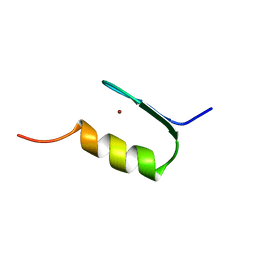 | | SWI5 ZINC FINGER DOMAIN 2, NMR, 45 STRUCTURES | | Descriptor: | SWI5, ZINC ION | | Authors: | Neuhaus, D, Nakaseko, Y, Schwabe, J.W.R, Rhodes, D, Klug, A. | | Deposit date: | 1996-04-04 | | Release date: | 1996-10-14 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two zinc-finger domains from SWI5 obtained using two-dimensional 1H nuclear magnetic resonance spectroscopy. A zinc-finger structure with a third strand of beta-sheet.
J.Mol.Biol., 228, 1992
|
|
2N8A
 
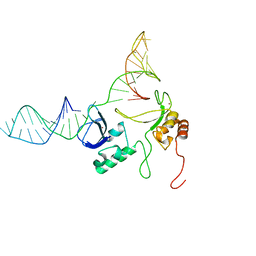 | | 1H, 13C and 15N chemical shift assignments and solution structure for PARP-1 F1F2 domains in complex with a DNA single-strand break | | Descriptor: | DNA (45-MER), Poly [ADP-ribose] polymerase 1, ZINC ION | | Authors: | Neuhaus, D, Eustermann, S, Yang, J, Wu, W. | | Deposit date: | 2015-10-08 | | Release date: | 2015-12-02 | | Last modified: | 2016-10-19 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Detection and Signaling of DNA Single-Strand Breaks by Human PARP-1.
Mol.Cell, 60, 2015
|
|
5NR6
 
 | |
5NR5
 
 | |
2L31
 
 | | Human PARP-1 zinc finger 2 | | Descriptor: | Poly [ADP-ribose] polymerase 1, ZINC ION | | Authors: | Neuhaus, D, Eustermann, S, Yang, J, Videler, H. | | Deposit date: | 2010-08-30 | | Release date: | 2011-02-02 | | Last modified: | 2020-02-26 | | Method: | SOLUTION NMR | | Cite: | The DNA-binding domain of human PARP-1 interacts with DNA single-strand breaks as a monomer through its second zinc finger.
J.Mol.Biol., 407, 2011
|
|
2L30
 
 | | Human PARP-1 zinc finger 1 | | Descriptor: | Poly [ADP-ribose] polymerase 1, ZINC ION | | Authors: | Neuhaus, D, Eustermann, S, Yang, J, Videler, H. | | Deposit date: | 2010-08-30 | | Release date: | 2011-02-02 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | The DNA-binding domain of human PARP-1 interacts with DNA single-strand breaks as a monomer through its second zinc finger.
J.Mol.Biol., 407, 2011
|
|
2KQC
 
 | | Second PBZ domain of human APLF protein | | Descriptor: | Aprataxin and PNK-like factor, ZINC ION | | Authors: | Neuhaus, D, Eustermann, S, Brockmann, C, Yang, J. | | Deposit date: | 2009-11-04 | | Release date: | 2010-01-19 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose).
Nat.Struct.Mol.Biol., 17, 2010
|
|
2KQB
 
 | | First PBZ domain of human APLF protein | | Descriptor: | Aprataxin and PNK-like factor, ZINC ION | | Authors: | Neuhaus, D, Eustermann, S, Brockmann, C, Yang, J. | | Deposit date: | 2009-11-04 | | Release date: | 2010-01-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose).
Nat.Struct.Mol.Biol., 17, 2010
|
|
2KQD
 
 | | First PBZ domain of human APLF protein in complex with ribofuranosyladenosine | | Descriptor: | ADENOSINE, Aprataxin and PNK-like factor, ZINC ION, ... | | Authors: | Neuhaus, D, Eustermann, S, Brockmann, C, Yang, J. | | Deposit date: | 2009-11-04 | | Release date: | 2010-01-19 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose).
Nat.Struct.Mol.Biol., 17, 2010
|
|
2KQE
 
 | | Second PBZ domain of human APLF protein in complex with ribofuranosyladenosine | | Descriptor: | ADENOSINE, Aprataxin and PNK-like factor, ZINC ION, ... | | Authors: | Neuhaus, D, Eustermann, S, Brockmann, C, Yang, J. | | Deposit date: | 2009-11-04 | | Release date: | 2010-01-19 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose).
Nat.Struct.Mol.Biol., 17, 2010
|
|
2LD1
 
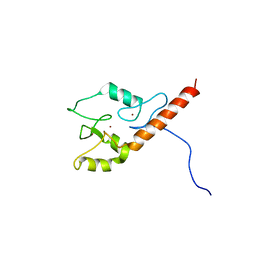 | |
1AUD
 
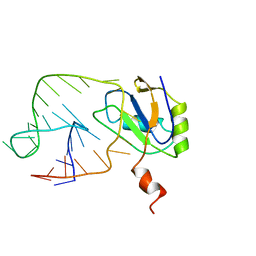 | | U1A-UTRRNA, NMR, 31 STRUCTURES | | Descriptor: | RNA 3UTR, U1A 102 | | Authors: | Allain, F.H.-T, Gubser, C.C, Howe, P.W.A, Nagai, K, Neuhaus, D, Varani, G. | | Deposit date: | 1997-08-22 | | Release date: | 1998-02-25 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Structural basis of the RNA-binding specificity of human U1A protein.
EMBO J., 16, 1997
|
|
1CDR
 
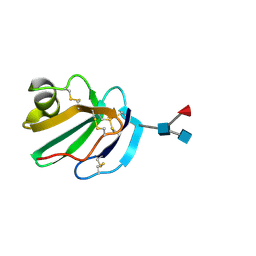 | | STRUCTURE OF A SOLUBLE, GLYCOSYLATED FORM OF THE HUMAN COMPLEMENT REGULATORY PROTEIN CD59 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CD59 | | Authors: | Fletcher, C.M, Harrison, R.A, Lachmann, P.J, Neuhaus, D. | | Deposit date: | 1994-06-01 | | Release date: | 1994-09-30 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Structure of a soluble, glycosylated form of the human complement regulatory protein CD59.
Structure, 2, 1994
|
|
1CDS
 
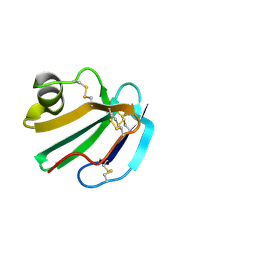 | | STRUCTURE OF A SOLUBLE, GLYCOSYLATED FORM OF THE HUMAN COMPLEMENT REGULATORY PROTEIN CD59 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD59 | | Authors: | Fletcher, C.M, Harrison, R.A, Lachmann, P.J, Neuhaus, D. | | Deposit date: | 1994-06-01 | | Release date: | 1994-09-30 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Structure of a soluble, glycosylated form of the human complement regulatory protein CD59.
Structure, 2, 1994
|
|
1NCS
 
 | | NMR STUDY OF SWI5 ZINC FINGER DOMAIN 1 | | Descriptor: | TRANSCRIPTIONAL FACTOR SWI5, ZINC ION | | Authors: | Dutnall, R.N, Neuhaus, D, Rhodes, D. | | Deposit date: | 1996-02-26 | | Release date: | 1996-06-10 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the first zinc finger domain of SWI5: a novel structural extension to a common fold.
Structure, 4, 1996
|
|
1HCP
 
 | |
7BBD
 
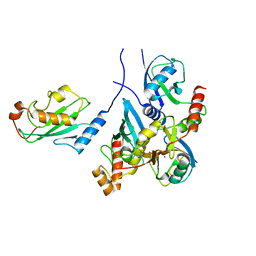 | | Crystal structure of monoubiquitinated TRIM21 RING (Ub-RING) In complex with ubiquitin charged Ube2N (Ube2N~Ub) and Ube2V2 | | Descriptor: | Polyubiquitin-B,E3 ubiquitin-protein ligase TRIM21, Polyubiquitin-C, Ubiquitin-conjugating enzyme E2 N, ... | | Authors: | Kiss, L, Neuhaus, D, James, L.C. | | Deposit date: | 2020-12-17 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | RING domains act as both substrate and enzyme in a catalytic arrangement to drive self-anchored ubiquitination.
Nat Commun, 12, 2021
|
|
7BBF
 
 | |
3MSP
 
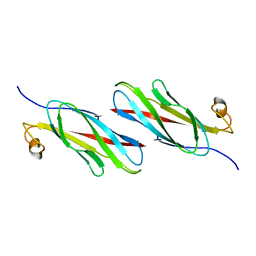 | | MOTILE MAJOR SPERM PROTEIN (MSP) OF ASCARIS SUUM, NMR, 20 STRUCTURES | | Descriptor: | MAJOR SPERM PROTEIN | | Authors: | Haaf, A, Leclaire III, L, Roberts, G, Kent, H.M, Roberts, T.M, Stewart, M, Neuhaus, D. | | Deposit date: | 1998-09-10 | | Release date: | 1999-04-20 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the motile major sperm protein (MSP) of Ascaris suum - evidence for two manganese binding sites and the possible role of divalent cations in filament formation.
J.Mol.Biol., 284, 1998
|
|
1E7J
 
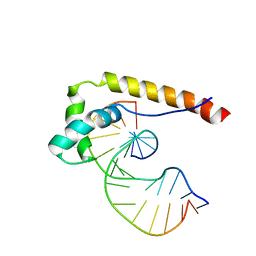 | | HMG-D complexed to a bulge DNA | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*AP*TP*TP*AP*AP*GP*AP*GP*CP*C)-3'), DNA (5'-D(*GP*GP*CP*TP*CP*AP*AP*TP*AP*TP*CP*G)-3'), HIGH MOBILITY GROUP PROTEIN D | | Authors: | Cerdan, R, Payet, D, Yang, J.-C, Travers, A.A, Neuhaus, D. | | Deposit date: | 2000-08-29 | | Release date: | 2001-03-05 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Hmg-D Complexed to a Bulge DNA: An NMR Model
Protein Sci., 10, 2001
|
|
2MY1
 
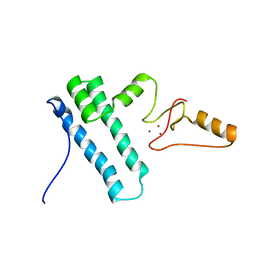 | | Solution structure of Bud31p | | Descriptor: | Pre-mRNA-splicing factor BUD31, ZINC ION | | Authors: | van Roon, A.M, Yang, J, Mathieu, D, Bermel, W, Nagai, K, Neuhaus, D. | | Deposit date: | 2015-01-19 | | Release date: | 2015-03-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | (113) Cd NMR Experiments Reveal an Unusual Metal Cluster in the Solution Structure of the Yeast Splicing Protein Bud31p.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
1CDQ
 
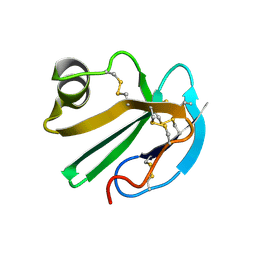 | | STRUCTURE OF A SOLUBLE, GLYCOSYLATED FORM OF THE HUMAN COMPLEMENT REGULATORY PROTEIN CD59 | | Descriptor: | CD59 | | Authors: | Fletcher, C.M, Harrison, R.A, Lachmann, P.J, Neuhaus, D. | | Deposit date: | 1994-06-01 | | Release date: | 1994-09-30 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structure of a soluble, glycosylated form of the human complement regulatory protein CD59.
Structure, 2, 1994
|
|
2BO5
 
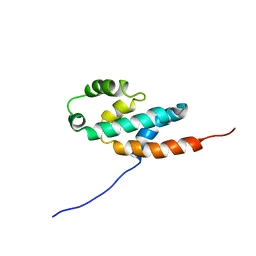 | | Bovine oligomycin sensitivity conferral protein N-terminal domain | | Descriptor: | ATP SYNTHASE OLIGOMYCIN SENSITIVITY CONFERRAL PROTEIN | | Authors: | Carbajo, R.J, Kellas, F.A, Runswick, M.J, Montgomery, M.G, Walker, J.E, Neuhaus, D. | | Deposit date: | 2005-04-07 | | Release date: | 2005-08-17 | | Last modified: | 2018-05-09 | | Method: | SOLUTION NMR | | Cite: | Structure of the F1-binding domain of the stator of bovine F1Fo-ATPase and how it binds an alpha-subunit.
J. Mol. Biol., 351, 2005
|
|
2BN6
 
 | | P-Element Somatic Inhibitor Protein | | Descriptor: | PSI | | Authors: | Ignjatovic, T, Yang, J.C, Butler, P.J.G, Neuhaus, D, Nagai, K. | | Deposit date: | 2005-03-21 | | Release date: | 2005-07-06 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of the Interaction between P-Element Somatic Inhibitor and U1-70K Essential for the Alternative Splicing of P-Element Transposase.
J.Mol.Biol., 351, 2005
|
|
2BN5
 
 | | P-Element Somatic Inhibitor Protein Complex with U1-70k proline-rich peptide | | Descriptor: | PSI, U1 SMALL NUCLEAR RIBONUCLEOPROTEIN 70 KDA | | Authors: | Ignjatovic, T, Yang, J.-C, Butler, P.J.G, Neuhaus, D, Nagai, K. | | Deposit date: | 2005-03-21 | | Release date: | 2005-07-06 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of the Interaction between P-Element Somatic Inhibitor and U1-70K Essential for the Alternative Splicing of P-Element Transposase.
J.Mol.Biol., 351, 2005
|
|
