4Z9E
 
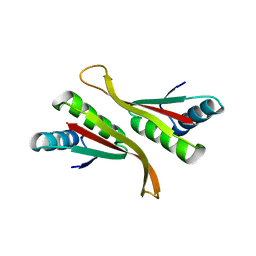 | | Alba from Thermoplasma volcanium | | Descriptor: | DNA/RNA-binding protein Alba | | Authors: | Ma, C, Lee, S.J, Pathak, C, Lee, B.J. | | Deposit date: | 2015-04-10 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Alba from Thermoplasma volcanium belongs to alpha-NAT's: An insight into the structural aspects of Tv Alba and its acetylation by Tv Ard1.
Arch.Biochem.Biophys., 590, 2016
|
|
5H4P
 
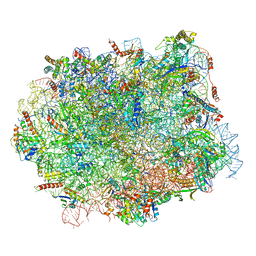 | | Structural snapshot of cytoplasmic pre-60S ribosomal particles bound with Nmd3, Lsg1, Tif6 and Reh1 | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Ma, C, Wu, S, Li, N, Chen, Y, Yan, K, Li, Z, Zheng, L, Lei, J, Woolford, J.L, Gao, N. | | Deposit date: | 2016-11-01 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural snapshot of cytoplasmic pre-60S ribosomal particles bound by Nmd3, Lsg1, Tif6 and Reh1
Nat. Struct. Mol. Biol., 24, 2017
|
|
5H5U
 
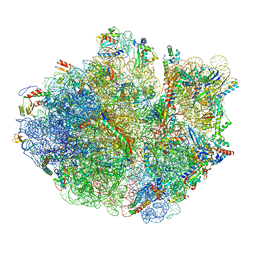 | | Mechanistic insights into the alternative translation termination by ArfA and RF2 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Ma, C, Kurita, D, Li, N, Chen, Y, Himeno, H, Gao, N. | | Deposit date: | 2016-11-09 | | Release date: | 2017-01-25 | | Last modified: | 2018-10-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanistic insights into the alternative translation termination by ArfA and RF2
Nature, 541, 2017
|
|
4PV6
 
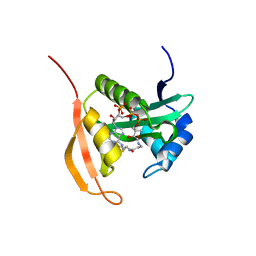 | | Crystal Structure Analysis of Ard1 from Thermoplasma volcanium | | Descriptor: | ACETYL COENZYME *A, COENZYME A, N-terminal acetyltransferase complex subunit [ARD1] | | Authors: | Ma, C, Lee, S.J, Lee, B.J. | | Deposit date: | 2014-03-15 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Structure of Thermoplasma volcanium Ard1 belongs to N-acetyltransferase family member suggesting multiple ligand binding modes with acetyl coenzyme A and coenzyme A.
Biochim.Biophys.Acta, 1844, 2014
|
|
6K6U
 
 | |
4PQP
 
 | | Crystal structure of human SNX14 PX domain in space group P43212 | | Descriptor: | GLYCEROL, Sorting nexin-14 | | Authors: | Mas, C, Norwood, S, Bugarcic, A, Kinna, G, Leneva, N, Kovtun, O, Teasdale, R, Collins, B. | | Deposit date: | 2014-03-03 | | Release date: | 2014-09-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Different Phosphoinositide Specificities of the PX Domains of Sorting Nexins Regulating G-protein Signaling.
J.Biol.Chem., 289, 2014
|
|
4PQO
 
 | | Structure of the human SNX14 PX domain in space group I41 | | Descriptor: | Sorting nexin-14 | | Authors: | Mas, C, Norwood, S, Bugarcic, A, Kinna, G, Leneva, N, Kovtun, O, Teasdale, R, Collins, B. | | Deposit date: | 2014-03-03 | | Release date: | 2014-09-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis for Different Phosphoinositide Specificities of the PX Domains of Sorting Nexins Regulating G-protein Signaling.
J.Biol.Chem., 289, 2014
|
|
2L2I
 
 | |
6WTT
 
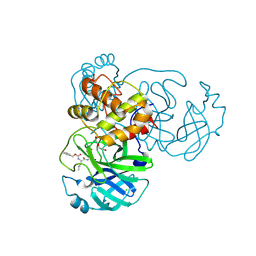 | | Crystals Structure of the SARS-CoV-2 (COVID-19) main protease with inhibitor GC-376 | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Sacco, M, Ma, C, Chen, Y, Wang, J. | | Deposit date: | 2020-05-03 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease.
Cell Res., 30, 2020
|
|
2KI2
 
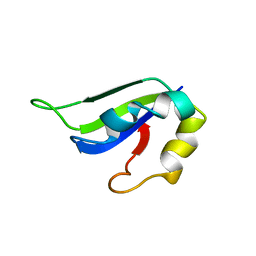 | | Solution Structure of ss-DNA Binding Protein 12RNP2 Precursor, HP0827(O25501_HELPY) form Helicobacter pylori | | Descriptor: | Ss-DNA binding protein 12RNP2 | | Authors: | Ma, C, Lee, J, Kim, J, Park, S, Kwon, A, Lee, B. | | Deposit date: | 2009-04-20 | | Release date: | 2009-10-20 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of HP0827 (O25501_HELPY) from Helicobacter pylori: model of the possible RNA-binding site
J.Biochem., 146, 2009
|
|
7JRN
 
 | | Crystal structure of the wild type SARS-CoV-2 papain-like protease (PLPro) with inhibitor GRL0617 | | Descriptor: | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, Non-structural protein 3, SULFATE ION, ... | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-08-12 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery of SARS-CoV-2 Papain-like Protease Inhibitors through a Combination of High-Throughput Screening and a FlipGFP-Based Reporter Assay.
Acs Cent.Sci., 7, 2021
|
|
2XA8
 
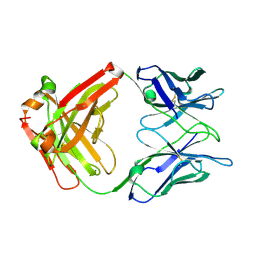 | | Crystal structure of the Fab domain of omalizumab at 2.41A | | Descriptor: | OMALIZUMAB HEAVY CHAIN, OMALIZUMAB LIGHT CHAIN | | Authors: | Huang, C.H, Hung, F.H.A, Lim, C, Chang, T.W, Ma, C. | | Deposit date: | 2010-03-30 | | Release date: | 2011-05-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural and Physical Basis for Anti-IgE Therapy.
Sci Rep, 5, 2015
|
|
2OAZ
 
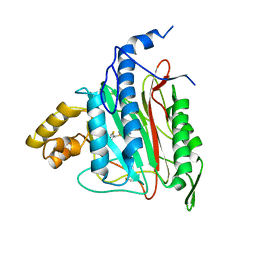 | | Human Methionine Aminopeptidase-2 Complexed with SB-587094 | | Descriptor: | COBALT (II) ION, N-(2-ISOPROPYLPHENYL)-3-[(2-THIENYLMETHYL)THIO]-1H-1,2,4-TRIAZOL-5-AMINE, human Methionine Amino Peptidase 2 | | Authors: | Marino Jr, J.P, Fisher, P.W, Hofmann, G.A, Kirkpatrick, R, Janson, C.A, Johnson, R.K, Ma, C, Mattern, M, Meek, T.D, Ryan, D, Schulz, C, Smith, W.W, Tew, D.G, Tomazek Jr, T.A, Veber, D.F, Xiong, W.C, Yamamoto, Y, Yamashita, K, Yang, G, Thompson, S.K. | | Deposit date: | 2006-12-18 | | Release date: | 2007-06-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Highly potent inhibitors of methionine aminopeptidase-2 based on a 1,2,4-triazole pharmacophore.
J.Med.Chem., 50, 2007
|
|
1ZXO
 
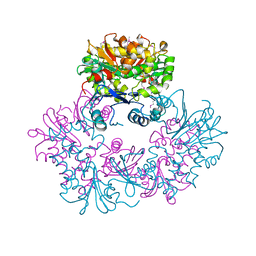 | | X-ray Crystal Structure of Protein Q8A1P1 from Bacteroides thetaiotaomicron. Northeast Structural Genomics Consortium Target BtR25. | | Descriptor: | conserved hypothetical protein Q8A1P1 | | Authors: | Kuzin, A.P, Yong, W, Forouhar, F, Vorobiev, S, Xiao, R, Ma, C, Acton, T, Montelione, G.T, Hunt, J, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-06-08 | | Release date: | 2005-07-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-Ray structure of the hypothetical protein Q8A1P1 at the resolution 3.2A. Northeast Structural Genomics Consortium target BtR25.
To be Published
|
|
2AJA
 
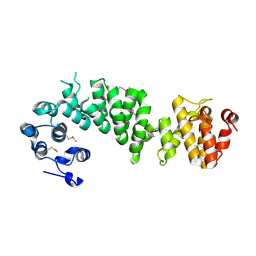 | | X-Ray structure of an ankyrin repeat family protein Q5ZSV0 from Legionella pneumophila. Northeast Structural Genomics Consortium target LgR21. | | Descriptor: | ankyrin repeat family protein | | Authors: | Kuzin, A.P, Chen, Y, Acton, T, Xiao, R, Conover, K, Ma, C, Kellie, R, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-08-01 | | Release date: | 2005-08-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-Ray structure of an ankyrin repeat family protein Q5ZSV0 from Legionella pneumophila.
To be Published
|
|
7PEG
 
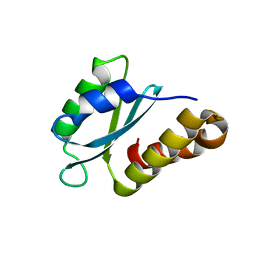 | | Structure of the sporulation/germination protein YhcN from Bacillus subtilis | | Descriptor: | Probable spore germination lipoprotein YhcN | | Authors: | Liu, B, Chan, H, Bauda, E, Contreras-Martel, C, Bellard, L, Villard, A.M, Mas, C, Neumann, E, Fenel, D, Favier, A, Serrano, M, Henriques, A.O.H, Rodrigues, C.D.A, Morlot, C. | | Deposit date: | 2021-08-10 | | Release date: | 2022-08-24 | | Last modified: | 2022-09-07 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural insights into ring-building motif domains involved in bacterial sporulation.
J.Struct.Biol., 214, 2022
|
|
3PFT
 
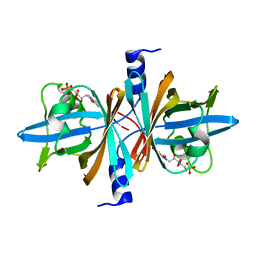 | | Crystal Structure of Untagged C54A Mutant Flavin Reductase (DszD) in Complex with FMN From Mycobacterium goodii | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavin reductase | | Authors: | Li, Q, Xu, P, Ma, C, Gu, L, Liu, X, Zhang, C, Li, N, Su, J, Li, B, Liu, S. | | Deposit date: | 2010-10-29 | | Release date: | 2011-11-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | The flavin reductase DSZD from a desulfurizing mycobacterium goodii strain: systemic manipulation and investigation based on the crystal structure
To be Published
|
|
7CFM
 
 | | Cryo-EM structure of the P395-bound GPBAR-Gs complex | | Descriptor: | 2-(ethylamino)-6-[3-(4-propan-2-ylphenyl)propanoyl]-7,8-dihydro-5H-pyrido[4,3-d]pyrimidine-4-carboxamide, CHOLESTEROL, G-protein coupled bile acid receptor 1, ... | | Authors: | Yang, F, Mao, C, Guo, L, Lin, J, Ming, Q, Xiao, P, Wu, X, Shen, Q, Guo, S, Shen, D, Lu, R, Zhang, L, Huang, S, Ping, Y, Zhang, C, Ma, C, Zhang, K, Liang, X, Shen, Y, Nan, F, Yi, F, Luca, V, Zhou, J, Jiang, C, Sun, J, Xie, X, Yu, X, Zhang, Y. | | Deposit date: | 2020-06-27 | | Release date: | 2020-09-09 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of GPBAR activation and bile acid recognition.
Nature, 587, 2020
|
|
7CFN
 
 | | Cryo-EM structure of the INT-777-bound GPBAR-Gs complex | | Descriptor: | (2S,4R)-4-[(3R,5S,6R,7R,8R,9S,10S,12S,13R,14S,17R)-6-ethyl-10,13-dimethyl-3,7,12-tris(oxidanyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]-2-methyl-pentanoic acid, CHOLESTEROL, G-protein coupled bile acid receptor 1, ... | | Authors: | Yang, F, Mao, C, Guo, L, Lin, J, Ming, Q, Xiao, P, Wu, X, Shen, Q, Guo, S, Shen, D, Lu, R, Zhang, L, Huang, S, Ping, Y, Zhang, C, Ma, C, Zhang, K, Liang, X, Shen, Y, Nan, F, Yi, F, Luca, V, Zhou, J, Jiang, C, Sun, J, Xie, X, Yu, X, Zhang, Y. | | Deposit date: | 2020-06-27 | | Release date: | 2020-09-09 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of GPBAR activation and bile acid recognition.
Nature, 587, 2020
|
|
4Y21
 
 | | Crystal Structure of Munc13-1 MUN domain | | Descriptor: | Protein unc-13 homolog A | | Authors: | Yang, X.Y, Wang, S, Sheng, Y, Zhang, M, Zou, W.J, Wu, L.J, Kang, L.J, Rizo, J, Zhang, R.G, Xu, T, Ma, C. | | Deposit date: | 2015-02-09 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Syntaxin opening by the MUN domain underlies the function of Munc13 in synaptic-vesicle priming.
Nat.Struct.Mol.Biol., 22, 2015
|
|
6GZ8
 
 | | First GerMN domain of the sporulation protein GerM from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, Spore germination protein GerM | | Authors: | Trouve, J, Mohamed, A, Leisico, F, Contreras-Martel, C, Liu, B, Mas, C, Rudner, D.Z, Rodrigues, C.D.A, Morlot, C. | | Deposit date: | 2018-07-03 | | Release date: | 2018-10-10 | | Last modified: | 2018-12-26 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural characterization of the sporulation protein GerM from Bacillus subtilis.
J. Struct. Biol., 204, 2018
|
|
6GZB
 
 | | Tandem GerMN domains of the sporulation protein GerM from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Spore germination protein GerM | | Authors: | Trouve, J, Mohamed, A, Leisico, F, Contreras-Martel, C, Liu, B, Mas, C, Rudner, D.Z, Rodrigues, C.D.A, Morlot, C. | | Deposit date: | 2018-07-03 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of the sporulation protein GerM from Bacillus subtilis.
J. Struct. Biol., 204, 2018
|
|
5GSQ
 
 | | Crystal structure of IgG Fc with a homogeneous glycoform and Antibody-Dependent Cellular Cytotoxicity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, C.-L, Hsu, J.-C, Lin, C.-W, Wu, C.-Y, Wong, C.-H, Ma, C. | | Deposit date: | 2016-08-17 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of a Homogeneous IgG-Fc Glycoform with the N-Glycan Designed to Maximize the Antibody Dependent Cellular Cytotoxicity
ACS Chem. Biol., 12, 2017
|
|
7CN9
 
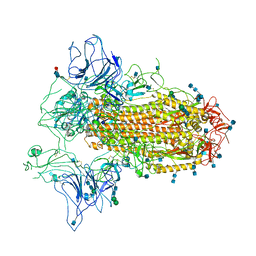 | | Cryo-EM structure of SARS-CoV-2 Spike ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ho, M, Chang, Y, Wang, C, Wu, Y, Huang, H, Chen, T, Lo, J.M, Chen, X, Ma, C. | | Deposit date: | 2020-07-30 | | Release date: | 2020-08-26 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | A Carbohydrate-Binding Protein from the Edible Lablab Beans Effectively Blocks the Infections of Influenza Viruses and SARS-CoV-2.
Cell Rep, 32, 2020
|
|
2K2U
 
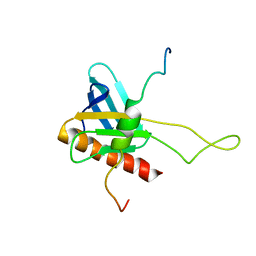 | | NMR Structure of the complex between Tfb1 subunit of TFIIH and the activation domain of VP16 | | Descriptor: | Alpha trans-inducing protein, RNA polymerase II transcription factor B subunit 1 | | Authors: | Langlois, C, Mas, C, Di Lello, P, Miller Jenkins, P.M, Legault, J, Omichinski, J.G. | | Deposit date: | 2008-04-11 | | Release date: | 2008-08-12 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Complex between the Tfb1 Subunit of TFIIH and the Activation Domain of VP16: Structural Similarities between VP16 and p53.
J.Am.Chem.Soc., 130, 2008
|
|
