1OFI
 
 | | Asymmetric complex between HslV and I-domain deleted HslU (H. influenzae) | | Descriptor: | 4-IODO-3-NITROPHENYL ACETYL-LEUCINYL-LEUCINYL-LEUCINYL-VINYLSULFONE, ADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ... | | Authors: | Kwon, A.R, Kessler, B.M, Overkleeft, H.S, McKay, D.B. | | Deposit date: | 2003-04-14 | | Release date: | 2003-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and Reactivity of an Asymmetric Complex between Hslv and I-Domain Deleted Hslu, a Prokaryotic Homolog of the Eukaryotic Proteasome
J.Mol.Biol., 330, 2003
|
|
1OFH
 
 | | Asymmetric complex between HslV and I-domain deleted HslU (H. influenzae) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV, ... | | Authors: | Kwon, A.R, Kessler, B.M, Overkleeft, H.S, McKay, D.B. | | Deposit date: | 2003-04-14 | | Release date: | 2003-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Reactivity of an Asymmetric Complex between Hslv and I-Domain Deleted Hslu, a Prokaryotic Homolog of the Eukaryotic Proteasome
J.Mol.Biol., 330, 2003
|
|
3FX7
 
 | |
3UI3
 
 | |
4Y1E
 
 | | SAV1875-C105D | | Descriptor: | Uncharacterized protein SAV1875 | | Authors: | Kim, H.J, Kwon, A.R, Lee, B.J. | | Deposit date: | 2015-02-07 | | Release date: | 2016-01-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional insight into the different oxidation states of SAV1875 from Staphylococcus aureus
Biochem.J., 473, 2016
|
|
4Y0N
 
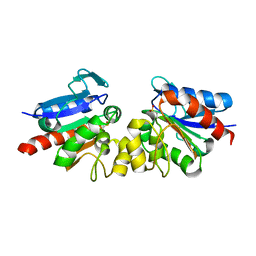 | | SAV1875 | | Descriptor: | Uncharacterized protein SAV1875 | | Authors: | Kim, H.J, Kwon, A.R, Lee, B.J. | | Deposit date: | 2015-02-06 | | Release date: | 2016-01-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional insight into the different oxidation states of SAV1875 from Staphylococcus aureus
Biochem.J., 473, 2016
|
|
4Y1F
 
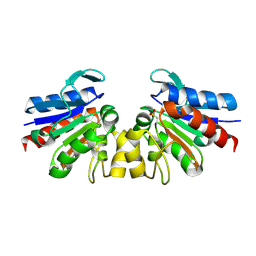 | | SAV1875-E17D | | Descriptor: | Uncharacterized protein SAV1875 | | Authors: | Kim, H.J, Kwon, A.R, Lee, B.J. | | Deposit date: | 2015-02-07 | | Release date: | 2016-01-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional insight into the different oxidation states of SAV1875 from Staphylococcus aureus
Biochem.J., 473, 2016
|
|
4Y1G
 
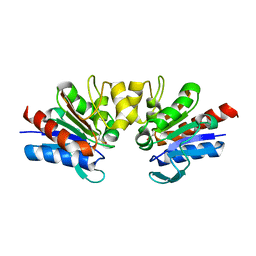 | | SAV1875-E17N | | Descriptor: | Uncharacterized protein SAV1875 | | Authors: | Kim, H.J, Kwon, A.R, Lee, B.J. | | Deposit date: | 2015-02-07 | | Release date: | 2016-01-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional insight into the different oxidation states of SAV1875 from Staphylococcus aureus
Biochem.J., 473, 2016
|
|
4Y1R
 
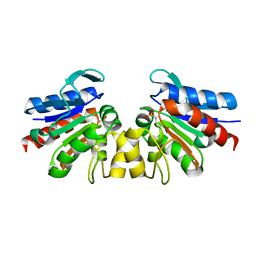 | | SAV1875-cysteinesulfonic acid | | Descriptor: | Uncharacterized protein SAV1875 | | Authors: | Kim, H.J, Kwon, A.R, Lee, B.J. | | Deposit date: | 2015-02-08 | | Release date: | 2016-01-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and functional insight into the different oxidation states of SAV1875 from Staphylococcus aureus
Biochem.J., 473, 2016
|
|
2KI2
 
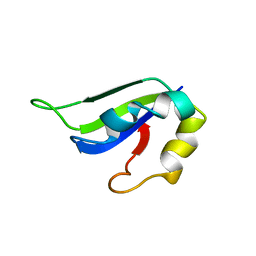 | | Solution Structure of ss-DNA Binding Protein 12RNP2 Precursor, HP0827(O25501_HELPY) form Helicobacter pylori | | Descriptor: | Ss-DNA binding protein 12RNP2 | | Authors: | Ma, C, Lee, J, Kim, J, Park, S, Kwon, A, Lee, B. | | Deposit date: | 2009-04-20 | | Release date: | 2009-10-20 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of HP0827 (O25501_HELPY) from Helicobacter pylori: model of the possible RNA-binding site
J.Biochem., 146, 2009
|
|
1WOG
 
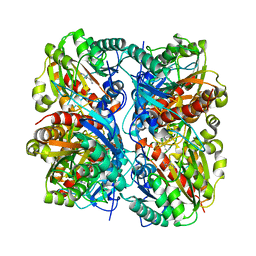 | | Crystal Structure of Agmatinase Reveals Structural Conservation and Inhibition Mechanism of the Ureohydrolase Superfamily | | Descriptor: | HEXANE-1,6-DIAMINE, MANGANESE (II) ION, agmatinase | | Authors: | Ahn, H.J, Kim, K.H, Lee, J, Ha, J.-Y, Lee, H.H, Kim, D, Yoon, H.-J, Kwon, A.-R, Suh, S.W. | | Deposit date: | 2004-08-18 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of agmatinase reveals structural conservation and inhibition mechanism of the ureohydrolase superfamily
J.Biol.Chem., 279, 2004
|
|
1WOI
 
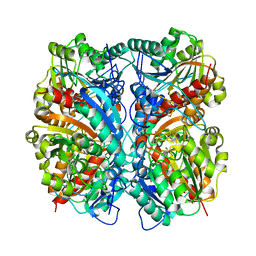 | | Crystal Structure of Agmatinase Reveals Structural Conservation and Inhibition Mechanism of the Ureohydrolase Superfamily | | Descriptor: | MANGANESE (II) ION, agmatinase | | Authors: | Ahn, H.J, Kim, K.H, Lee, J, Ha, J.-Y, Lee, H.H, Kim, D, Yoon, H.-J, Kwon, A.-R, Suh, S.W. | | Deposit date: | 2004-08-18 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of agmatinase reveals structural conservation and inhibition mechanism of the ureohydrolase superfamily
J.Biol.Chem., 279, 2004
|
|
3CNK
 
 | | Crystal Structure of the dimerization domain of human filamin A | | Descriptor: | Filamin-A, SULFATE ION | | Authors: | Lee, B.J, Seo, M.D, Seok, S.H, Lee, S.J, Kwon, A.R, Im, H. | | Deposit date: | 2008-03-26 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the dimerization domain of human filamin A
Proteins, 75, 2008
|
|
1WOH
 
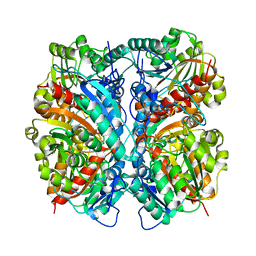 | | Crystal Structure of Agmatinase Reveals Structural Conservation and Inhibition Mechanism of the Ureohydrolase Superfamily | | Descriptor: | agmatinase | | Authors: | Ahn, H.J, Kim, K.H, Lee, J, Ha, J.-Y, Lee, H.H, Kim, D, Yoon, H.-J, Kwon, A.-R, Suh, S.W. | | Deposit date: | 2004-08-18 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of agmatinase reveals structural conservation and inhibition mechanism of the ureohydrolase superfamily
J.Biol.Chem., 279, 2004
|
|
6KHL
 
 | | Lipase (Blocked form) | | Descriptor: | (2~{R})-1-phenylpropan-2-ol, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kim, H.J, Kwon, A.R. | | Deposit date: | 2019-07-16 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Closed, blocked, and open states of lipase from type II Cutibacterium acnes
To Be Published
|
|
5XR2
 
 | | SAV0551 | | Descriptor: | LACTIC ACID, Protein/nucleic acid deglycase HchA, ZINC ION | | Authors: | Kim, H.J, Kwon, A.R, Lee, B.J. | | Deposit date: | 2017-06-07 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional studies of SAV0551 fromStaphylococcus aureusas a chaperone and glyoxalase III.
Biosci. Rep., 37, 2017
|
|
5XR3
 
 | | SAV0551 with glyoxylate | | Descriptor: | GLYOXYLIC ACID, Protein/nucleic acid deglycase HchA | | Authors: | Kim, H.J, Kwon, A.R, Lee, B.J. | | Deposit date: | 2017-06-08 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural and functional studies of SAV0551 fromStaphylococcus aureusas a chaperone and glyoxalase III.
Biosci. Rep., 37, 2017
|
|
6KHK
 
 | | Lipase (Closed form) | | Descriptor: | GLYCEROL, Hydrolase, alpha/beta domain protein | | Authors: | Kim, H.J, Kwon, A.R. | | Deposit date: | 2019-07-16 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Closed, blocked, and open states of lysophospholipase from type II Cutibacterium acnes
To Be Published
|
|
5YU2
 
 | | Structure of Ribonuclease YabJ | | Descriptor: | Translation initiation inhibitor homologue | | Authors: | Kim, H.J, Kwon, A.R, Lee, B.J. | | Deposit date: | 2017-11-20 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A novel chlorination-induced ribonuclease YabJ fromStaphylococcus aureus.
Biosci. Rep., 38, 2018
|
|
6KHM
 
 | | Lipase (Open form) | | Descriptor: | Hydrolase, alpha/beta domain protein, heptane-1,1-diol, ... | | Authors: | Kim, H.J, Kwon, A.R. | | Deposit date: | 2019-07-16 | | Release date: | 2020-07-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Closed, blocked, and open states of lipase from type II Cutibacterium acnes
To Be Published
|
|
1UHE
 
 | | Crystal structure of aspartate decarboxylase, isoaspargine complex | | Descriptor: | Aspartate 1-decarboxylase alpha chain, Aspartate 1-decarboxylase beta chain, N~2~-(2-AMINO-1-METHYL-2-OXOETHYLIDENE)ASPARAGINATE | | Authors: | Lee, B.I, Kwon, A.-R, Han, B.W, Suh, S.W. | | Deposit date: | 2003-07-01 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the schiff base intermediate prior to decarboxylation in the catalytic cycle of aspartate alpha-decarboxylase
J.Mol.Biol., 340, 2004
|
|
1UHD
 
 | | Crystal structure of aspartate decarboxylase, pyruvoly group bound form | | Descriptor: | Aspartate 1-decarboxylase alpha chain, Aspartate 1-decarboxylase beta chain | | Authors: | Lee, B.I, Kwon, A.-R, Han, B.W, Suh, S.W. | | Deposit date: | 2003-07-01 | | Release date: | 2004-07-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the schiff base intermediate prior to decarboxylation in the catalytic cycle of aspartate alpha-decarboxylase
J.Mol.Biol., 340, 2004
|
|
