8IQ6
 
 | | Cryo-EM structure of Latanoprost-bound prostaglandin-F2-alpha receptor-miniGq-Nb35 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Lv, X, Gao, K, Nie, J, Zhang, X, Zhang, S, Ren, Y, Li, Q, Huang, J, Liu, L, Zhang, X, Sun, X, Zhang, W, Liu, X. | | Deposit date: | 2023-03-15 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of human prostaglandin F 2 alpha receptor reveal the mechanism of ligand and G protein selectivity.
Nat Commun, 14, 2023
|
|
8IQ4
 
 | | Cryo-EM structure of Carboprost-bound prostaglandin-F2-alpha receptor-miniGq-Nb35 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Lv, X, Gao, K, Nie, J, Zhang, X, Zhang, S, Ren, Y, Li, Q, Huang, J, Liu, L, Zhang, X, Sun, X, Zhang, W, Liu, X. | | Deposit date: | 2023-03-15 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures of human prostaglandin F 2 alpha receptor reveal the mechanism of ligand and G protein selectivity.
Nat Commun, 14, 2023
|
|
7CVQ
 
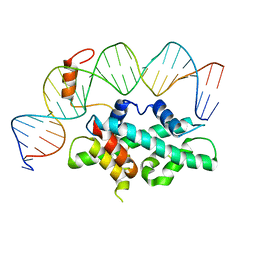 | | crystal structure of Arabidopsis CO CCT domain in complex with NF-YB2/YC3 and FT CORE1 DNA | | Descriptor: | Chimera of Nuclear transcription factor Y subunit C-3 and Zinc finger protein CONSTANS, FT CORE1 DNA forward strand, FT CORE1 DNA reverse strand, ... | | Authors: | Lv, X, Du, J. | | Deposit date: | 2020-08-26 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural insights into the multivalent binding of the Arabidopsis FLOWERING LOCUS T promoter by the CO-NF-Y master transcription factor complex.
Plant Cell, 33, 2021
|
|
7CVO
 
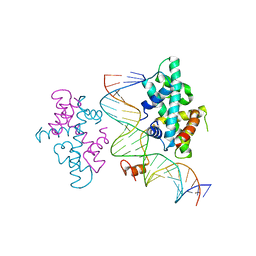 | | crystal structure of Arabidopsis CO CCT domain in complex with NF-YB3/YC4 and FT CORE2 DNA | | Descriptor: | Chimera of Nuclear transcription factor Y subunit C-4 and Zinc finger protein CONSTANS, FT CORE2 DNA forward strand, FT CORE2 DNA reverse strand, ... | | Authors: | Lv, X, Du, J. | | Deposit date: | 2020-08-26 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the multivalent binding of the Arabidopsis FLOWERING LOCUS T promoter by the CO-NF-Y master transcription factor complex.
Plant Cell, 33, 2021
|
|
5ZNP
 
 | | Crystal structure of PtSHL in complex with an H3K4me3 peptide | | Descriptor: | 15-mer peptide from Histone H3.2, SHORT LIFE family protein, ZINC ION | | Authors: | Lv, X, Du, J. | | Deposit date: | 2018-04-10 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dual recognition of H3K4me3 and H3K27me3 by a plant histone reader SHL.
Nat Commun, 9, 2018
|
|
5ZNR
 
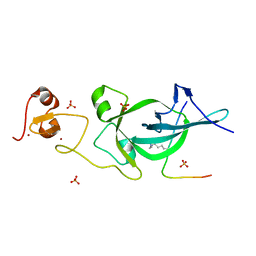 | | Crystal structure of PtSHL in complex with an H3K27me3 peptide | | Descriptor: | 17-mer peptide from Histone H3.2, SHORT LIFE family protein, SULFATE ION, ... | | Authors: | Lv, X, Du, J. | | Deposit date: | 2018-04-10 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Dual recognition of H3K4me3 and H3K27me3 by a plant histone reader SHL.
Nat Commun, 9, 2018
|
|
7VSP
 
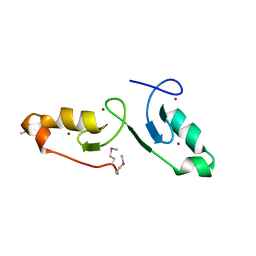 | | Crystal strcuture of the tandem B-Box domains of COL2 | | Descriptor: | ZINC ION, Zinc finger protein CONSTANS-LIKE 2 | | Authors: | Lv, X, Liu, R, Du, J. | | Deposit date: | 2021-10-27 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis of CONSTANS oligomerization in FLOWERING LOCUS T activation.
J Integr Plant Biol, 64, 2022
|
|
5JYY
 
 | | Structure-based Tetravalent Zanamivir with Potent Inhibitory Activity against Drug-resistant Influenza Viruses | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-2,6-anhydro-4-carbamimidamido-3,4,5-trideoxy-7-O-[(2-methoxyethyl)carbamoyl]-D-glycero-D-galacto-non-2-enon ic acid, CALCIUM ION, ... | | Authors: | Fu, L, Wu, Y, Bi, Y, Zhang, S, Lv, X, Qi, J, Li, Y, Lu, X, Yan, J, Gao, G.F, Li, X. | | Deposit date: | 2016-05-15 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Tetravalent Zanamivir with Potent Inhibitory Activity against Drug-Resistant Influenza Viruses
J.Med.Chem., 59, 2016
|
|
6KTR
 
 | | Crystal structure of fibroblast growth factor 19 in complex with Fab | | Descriptor: | Fibroblast growth factor 19, G1A8-Fab-HC, G1A8-Fab-LC, ... | | Authors: | Liu, H, Zheng, S, Hou, X, Liu, X, Lv, X, Li, Y, Li, W, Sui, J. | | Deposit date: | 2019-08-28 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.59775758 Å) | | Cite: | Novel Abs targeting the N-terminus of fibroblast growth factor 19 inhibit hepatocellular carcinoma growth without bile-acid-related side-effects.
Cancer Sci., 111, 2020
|
|
7VSQ
 
 | |
7E6Q
 
 | | Crystal structure of influenza A virus neuraminidase N5 complexed with 4'-phenyl-1,2,3-triazolylated oseltamivir carboxylate | | Descriptor: | (3R,4R,5S)-4-acetamido-3-pentan-3-yloxy-5-(4-phenyl-1,2,3-triazol-1-yl)cyclohexene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wang, P.F, Babayemi, O.O, Li, C.N, Fu, L.F, Zhang, S.S, Qi, J.X, Lv, X, Li, X.B. | | Deposit date: | 2021-02-23 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based design of 5'-substituted 1,2,3-triazolylated oseltamivir derivatives as potent influenza neuraminidase inhibitors.
Rsc Adv, 11, 2021
|
|
3BS8
 
 | | Crystal structure of Glutamate 1-Semialdehyde Aminotransferase complexed with pyridoxamine-5'-phosphate From Bacillus subtilis | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Glutamate-1-semialdehyde 2,1-aminomutase | | Authors: | Ge, H, Fan, J, Teng, M, Niu, L. | | Deposit date: | 2007-12-22 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Glutamate1-semialdehyde aminotransferase from Bacillus subtilis with bound pyridoxamine-5'-phosphate
Biochem.Biophys.Res.Commun., 402, 2010
|
|
6RJ3
 
 | | Crystal structure of PHGDH in complex with compound 15 | | Descriptor: | 4-[(1~{R})-1-[(2-methyl-5-phenyl-pyrazol-3-yl)carbonylamino]ethyl]benzoic acid, D-3-phosphoglycerate dehydrogenase, SULFATE ION | | Authors: | Bader, G, Wolkerstorfer, B, Zoephel, A. | | Deposit date: | 2019-04-26 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Intracellular Trapping of the Selective Phosphoglycerate Dehydrogenase (PHGDH) InhibitorBI-4924Disrupts Serine Biosynthesis.
J.Med.Chem., 62, 2019
|
|
6RJ6
 
 | | Crystal structure of PHGDH in complex with BI-4924 | | Descriptor: | 2-[4-[(1~{S})-1-[[4,5-bis(chloranyl)-1,6-dimethyl-indol-2-yl]carbonylamino]-2-oxidanyl-ethyl]phenyl]sulfonylethanoic acid, D-3-phosphoglycerate dehydrogenase | | Authors: | Bader, G, Wolkerstorfer, B, Zoephel, A. | | Deposit date: | 2019-04-26 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Intracellular Trapping of the Selective Phosphoglycerate Dehydrogenase (PHGDH) InhibitorBI-4924Disrupts Serine Biosynthesis.
J.Med.Chem., 62, 2019
|
|
6RIH
 
 | | Crystal structure of PHGDH in complex with compound 9 | | Descriptor: | D-3-phosphoglycerate dehydrogenase, SULFATE ION, ~{N}-cyclopropyl-2-methyl-5-phenyl-pyrazole-3-carboxamide | | Authors: | Bader, G, Wolkerstorfer, B, Zoephel, A. | | Deposit date: | 2019-04-24 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Intracellular Trapping of the Selective Phosphoglycerate Dehydrogenase (PHGDH) InhibitorBI-4924Disrupts Serine Biosynthesis.
J.Med.Chem., 62, 2019
|
|
6RJ2
 
 | | Crystal structure of PHGDH in complex with compound 40 | | Descriptor: | D-3-phosphoglycerate dehydrogenase, SULFATE ION, ~{N}-[(1~{R})-1-[4-(ethanoylsulfamoyl)phenyl]ethyl]-2-methyl-5-phenyl-pyrazole-3-carboxamide | | Authors: | Bader, G, Wolkerstorfer, B, Zoephel, A. | | Deposit date: | 2019-04-26 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Intracellular Trapping of the Selective Phosphoglycerate Dehydrogenase (PHGDH) InhibitorBI-4924Disrupts Serine Biosynthesis.
J.Med.Chem., 62, 2019
|
|
6RJ5
 
 | | Crystal structure of PHGDH in complex with compound 39 | | Descriptor: | 2-methyl-~{N}-[(1~{R})-1-[4-(methylsulfonylcarbamoyl)phenyl]ethyl]-5-phenyl-pyrazole-3-carboxamide, D-3-phosphoglycerate dehydrogenase, SULFATE ION | | Authors: | Bader, G, Wolkerstorfer, B, Zoephel, A. | | Deposit date: | 2019-04-26 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Intracellular Trapping of the Selective Phosphoglycerate Dehydrogenase (PHGDH) InhibitorBI-4924Disrupts Serine Biosynthesis.
J.Med.Chem., 62, 2019
|
|
6CWA
 
 | |
5Z8N
 
 | |
5Z8L
 
 | |
4NXF
 
 | | Crystal structure of iLOV-I486(2LT) at pH 8.0 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Wang, J, Liu, X, Li, J. | | Deposit date: | 2013-12-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.766 Å) | | Cite: | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
4NXE
 
 | | Crystal structure of iLOV-I486(2LT) at pH 6.5 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Wang, J, Liu, X, Li, J. | | Deposit date: | 2013-12-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
4NXB
 
 | | Crystal structure of iLOV-I486(2LT) at pH 7.0 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Wang, J, Li, J, Liu, X. | | Deposit date: | 2013-12-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.561 Å) | | Cite: | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
4NXG
 
 | | Crystal structure of iLOV-I486z(2LT) at pH 9.0 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Wang, J, Liu, X, Li, J. | | Deposit date: | 2013-12-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
4NX2
 
 | | Crystal structure of DCYRS complexed with DCY | | Descriptor: | 3,5-dichloro-L-tyrosine, Tyrosine--tRNA ligase | | Authors: | Wang, J, Gong, W, Li, J, Gao, F, Li, H. | | Deposit date: | 2013-12-08 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
