1RV1
 
 | | CRYSTAL STRUCTURE OF HUMAN MDM2 WITH AN IMIDAZOLINE INHIBITOR | | Descriptor: | CIS-[4,5-BIS-(4-BROMOPHENYL)-2-(2-ETHOXY-4-METHOXYPHENYL)-4,5-DIHYDROIMIDAZOL-1-YL]-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]METHANONE, Ubiquitin-protein ligase E3 Mdm2 | | Authors: | Lukacs, C, Kammlott, U, Graves, B. | | Deposit date: | 2003-12-12 | | Release date: | 2004-01-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | In vivo activation of the p53 pathway by small-molecule antagonists of MDM2.
Science, 303, 2004
|
|
4GFG
 
 | | Crystal structure of spleen tyrosine kinase complexed with r9021 | | Descriptor: | 6-{[(1R,2S)-2-aminocyclohexyl]amino}-4-[(5,6-dimethylpyridin-2-yl)amino]pyridazine-3-carboxamide, Tyrosine-protein kinase SYK | | Authors: | Lukacs, C, Slade, M. | | Deposit date: | 2012-08-03 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A specific SYK inhibitor blocks B Cell Receptor, Fc Receptor, and Toll-like Receptor 9 pathways for the treatment of inflammatory diseases.
To be Published
|
|
4J7D
 
 | | The 1.25A crystal structure of humanized Xenopus MDM2 with a nutlin fragment, RO5045331 | | Descriptor: | (4S,5R)-2-(4-tert-butyl-2-ethoxyphenyl)-4,5-bis(4-chlorophenyl)-4,5-dimethyl-4,5-dihydro-1H-imidazole, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Janson, C, Lukacs, C, Graves, B. | | Deposit date: | 2013-02-13 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Deconstruction of a nutlin: dissecting the binding determinants of a potent protein-protein interaction inhibitor.
ACS Med Chem Lett, 4, 2013
|
|
4J74
 
 | | The 1.2A crystal structure of humanized Xenopus MDM2 with RO0503918 - a nutlin fragment | | Descriptor: | (4S,5R)-4,5-bis(4-chlorophenyl)-2-methyl-4,5-dihydro-1H-imidazole, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Janson, C, Lukacs, C, Kammlott, U, Graves, B. | | Deposit date: | 2013-02-12 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Deconstruction of a nutlin: dissecting the binding determinants of a potent protein-protein interaction inhibitor.
ACS Med Chem Lett, 4, 2013
|
|
4J7E
 
 | | The 1.63A crystal structure of humanized Xenopus MDM2 with a nutlin fragment, RO5524529 | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, SULFATE ION, [(4S,5R)-4,5-bis(4-chlorophenyl)-2,4,5-trimethyl-4,5-dihydro-1H-imidazol-1-yl]{4-[3-(methylsulfonyl)propyl]piperazin-1-yl}methanone | | Authors: | Janson, C, Lukacs, C, Graves, B. | | Deposit date: | 2013-02-13 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Deconstruction of a nutlin: dissecting the binding determinants of a potent protein-protein interaction inhibitor.
ACS Med Chem Lett, 4, 2013
|
|
4IPF
 
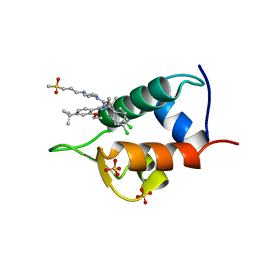 | | The 1.7A crystal structure of humanized Xenopus MDM2 with RO5045337 | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, SULFATE ION, [(4S,5R)-2-(4-tert-butyl-2-ethoxyphenyl)-4,5-bis(4-chlorophenyl)-4,5-dimethyl-4,5-dihydro-1H-imidazol-1-yl]{4-[3-(methylsulfonyl)propyl]piperazin-1-yl}methanone | | Authors: | Graves, B.J, Lukacs, C, Kammlott, R.U, Crowther, R. | | Deposit date: | 2013-01-09 | | Release date: | 2013-02-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | MDM2 Small-Molecule Antagonist RG7112 Activates p53 Signaling and Regresses Human Tumors in Preclinical Cancer Models.
Cancer Res., 73, 2013
|
|
4J44
 
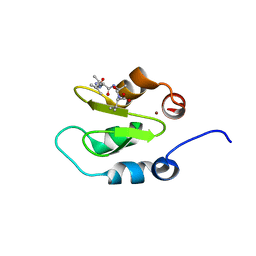 | | Crystal structure of XIAP-BIR2 domain with AIAV bound | | Descriptor: | E3 ubiquitin-protein ligase XIAP, PEPTIDE (ALA-ILE-ALA-VAL), ZINC ION | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2013-02-06 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structure of XIAP BIR2: understanding the selectivity of the BIR domains.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4J45
 
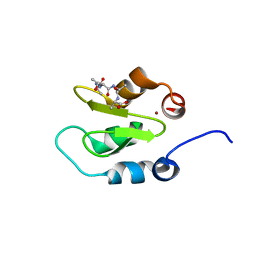 | | Crystal structure of XIAP-BIR2 domain with ATAA bound | | Descriptor: | E3 ubiquitin-protein ligase XIAP, PEPTIDE (ALA-THR-ALA-ALA), ZINC ION | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2013-02-06 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The structure of XIAP BIR2: understanding the selectivity of the BIR domains.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4J47
 
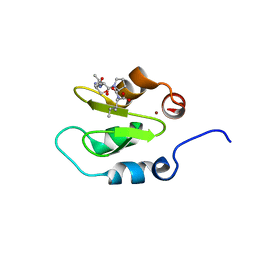 | | Crystal structure of XIAP-BIR2 domain with SVPI bound | | Descriptor: | E3 ubiquitin-protein ligase XIAP, PEPTIDE (SER-VAL-PRO-ILE), ZINC ION | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2013-02-06 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The structure of XIAP BIR2: understanding the selectivity of the BIR domains.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4J3Y
 
 | | Crystal structure of XIAP-BIR2 domain | | Descriptor: | E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2013-02-06 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The structure of XIAP BIR2: understanding the selectivity of the BIR domains.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4J46
 
 | | Crystal structure of XIAP-BIR2 domain with AVPI bound | | Descriptor: | E3 ubiquitin-protein ligase XIAP, PEPTIDE (ALA-VAL-PRO-ILE), ZINC ION | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2013-02-06 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The structure of XIAP BIR2: understanding the selectivity of the BIR domains.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4J48
 
 | | Crystal structure of XIAP-BIR2 domain with AMRV bound | | Descriptor: | E3 ubiquitin-protein ligase XIAP, GLYCEROL, PEPTIDE (ALA-MET-ARG-VAL), ... | | Authors: | Gosu, R. | | Deposit date: | 2013-02-06 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of XIAP BIR2: understanding the selectivity of the BIR domains.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3U15
 
 | | Structure of hDMX with Dimer Inducing Indolyl Hydantoin RO-2443 | | Descriptor: | (5Z)-5-[(6-chloro-7-methyl-1H-indol-3-yl)methylidene]-3-(3,4-difluorobenzyl)imidazolidine-2,4-dione, Protein Mdm4, SULFATE ION | | Authors: | Lukacs, C.M, Janson, C.A, Graves, B.J. | | Deposit date: | 2011-09-29 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Activation of the p53 pathway by small-molecule-induced MDM2 and MDMX dimerization.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3VBG
 
 | | Structure of hDM2 with Dimer Inducing Indolyl Hydantoin RO-2443 | | Descriptor: | (5Z)-5-[(6-chloro-7-methyl-1H-indol-3-yl)methylidene]-3-(3,4-difluorobenzyl)imidazolidine-2,4-dione, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Lukacs, C.M, Janson, C.A, Graves, B.J. | | Deposit date: | 2012-01-02 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Activation of the p53 pathway by small-molecule-induced MDM2 and MDMX dimerization.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ORN
 
 | | Mitogen-activated protein kinase kinase 1 (MEK1) in complex with CH4987655 and MgAMP-PNP | | Descriptor: | 3,4-difluoro-2-[(2-fluoro-4-iodophenyl)amino]-N-(2-hydroxyethoxy)-5-[(3-oxo-1,2-oxazinan-2-yl)methyl]benzamide, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Lukacs, C.M, Janson, C, Schuck, V, Belunis, C. | | Deposit date: | 2010-09-07 | | Release date: | 2011-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design and synthesis of novel allosteric MEK inhibitor CH4987655 as an orally available anticancer agent.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3OS3
 
 | | Mitogen-activated protein kinase kinase 1 (MEK1) in complex with CH4858061 and MgATP | | Descriptor: | 2-[(4-ethynyl-2-fluorophenyl)amino]-3,4-difluoro-N-(2-hydroxyethoxy)-5-{[(2-hydroxyethoxy)imino]methyl}benzamide, ADENOSINE-5'-TRIPHOSPHATE, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Lukacs, C.M, Janson, C, Schuck, V, Belunis, C. | | Deposit date: | 2010-09-08 | | Release date: | 2011-07-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design and synthesis of novel allosteric MEK inhibitor CH4987655 as an orally available anticancer agent.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3WIG
 
 | | Human MEK1 kinase in complex with CH5126766 and MgAMP-PNP | | Descriptor: | CHLORIDE ION, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Lukacs, C.M, Janson, C, Schuck, V. | | Deposit date: | 2013-09-12 | | Release date: | 2014-06-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Disruption of CRAF-Mediated MEK Activation Is Required for Effective MEK Inhibition in KRAS Mutant Tumors
Cancer Cell, 25, 2014
|
|
2FVD
 
 | | Cyclin Dependent Kinase 2 (CDK2) with diaminopyrimidine inhibitor | | Descriptor: | (4-AMINO-2-{[1-(METHYLSULFONYL)PIPERIDIN-4-YL]AMINO}PYRIMIDIN-5-YL)(2,3-DIFLUORO-6-METHOXYPHENYL)METHANONE, Cell division protein kinase 2 | | Authors: | Crowther, R.L, Lukacs, C.M, Kammlott, R.U. | | Deposit date: | 2006-01-30 | | Release date: | 2006-10-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of [4-Amino-2-(1-methanesulfonylpiperidin-4-ylamino)pyrimidin-5-yl](2,3-difluoro-6- methoxyphenyl)methanone (R547), a potent and selective cyclin-dependent kinase inhibitor with significant in vivo antitumor activity.
J.Med.Chem., 49, 2006
|
|
3LE6
 
 | | The structure of cyclin dependent kinase 2 (CKD2) with a pyrazolobenzodiazepine inhibitor | | Descriptor: | 5-(2-chlorophenyl)-3-methyl-7-nitropyrazolo[3,4-b][1,4]benzodiazepine, Cell division protein kinase 2 | | Authors: | Lukacs, C.M, Swain, A, Crowther, R.L, Kammlott, R.U, Liu, J.J. | | Deposit date: | 2010-01-14 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pyrazolobenzodiazepines: part I. Synthesis and SAR of a potent class of kinase inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5E0I
 
 | |
5C5R
 
 | | CRYSTAL STRUCTURE OF HUMAN TANKYRASE-2 IN COMPLEX WITH A PYRANOPYRIDONE INHIBITOR | | Descriptor: | (7R)-2-hydroxy-7-(propan-2-yl)-7,8-dihydro-5H-pyrano[4,3-b]pyridine-3-carbonitrile, SULFATE ION, Tankyrase-2, ... | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2015-06-21 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Fragment-Based Drug Design of Novel Pyranopyridones as Cell Active and Orally Bioavailable Tankyrase Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
5C5P
 
 | | CRYSTAL STRUCTURE OF HUMAN TANKYRASE-2 IN COMPLEX WITH A PYRANOPYRIDONE INHIBITOR | | Descriptor: | (3R)-3-(1-hydroxy-2-methylpropan-2-yl)-1,3,4,5-tetrahydro-6H-pyrano[4,3-c]isoquinolin-6-one, SULFATE ION, Tankyrase-2, ... | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2015-06-21 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Fragment-Based Drug Design of Novel Pyranopyridones as Cell Active and Orally Bioavailable Tankyrase Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
5C5Q
 
 | | CRYSTAL STRUCTURE OF HUMAN TANKYRASE-2 IN COMPLEX WITH A PYRANOPYRIDONE INHIBITOR | | Descriptor: | (3R)-10-methyl-3-(propan-2-yl)-1,3,4,5-tetrahydro-6H-pyrano[4,3-c]isoquinolin-6-one, SULFATE ION, Tankyrase-2, ... | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2015-06-21 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment-Based Drug Design of Novel Pyranopyridones as Cell Active and Orally Bioavailable Tankyrase Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
8GJS
 
 | | Stapled Peptide ALRN-6924 Bound to MDMX | | Descriptor: | ACE-LEU-THR-PHE-ALA-GLU-TYR-TRP-ALA-GLN-LEU-DAL-ALA-ALA-ALA-ALA-ALA-DAL, Protein Mdm4 | | Authors: | Graves, B.J, Janson, C, Lukacs, C. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Sulanemadlin (ALRN-6924), the First Cell-Permeating, Stabilized alpha-Helical Peptide in Clinical Development.
J.Med.Chem., 66, 2023
|
|
4MQ2
 
 | | The crystal structure of DYRK1a with a bound pyrido[2,3-d]pyrimidine inhibitor | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 1A, PENTAETHYLENE GLYCOL, SULFATE ION, ... | | Authors: | Lukacs, C.M, Janson, C.A, Garvie, C, Liang, L. | | Deposit date: | 2013-09-15 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Pyrido[2,3-d]pyrimidines: Discovery and preliminary SAR of a novel series of DYRK1B and DYRK1A inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
