1AKP
 
 | | SEQUENTIAL 1H,13C AND 15N NMR ASSIGNMENTS AND SOLUTION CONFORMATION OF APOKEDARCIDIN | | Descriptor: | APOKEDARCIDIN | | Authors: | Constantine, K.L, Colson, K.L, Wittekind, M, Friedrichs, M.S, Zein, N, Tuttle, J, Langley, D.R, Leet, J.E, Schroeder, D.R, Lam, K.S, Farmer II, B.T, Metzler, W.J, Bruccoleri, R.E, Mueller, L. | | Deposit date: | 1994-06-20 | | Release date: | 1994-08-31 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Sequential 1H, 13C, and 15N NMR assignments and solution conformation of apokedarcidin.
Biochemistry, 33, 1994
|
|
8SGE
 
 | | KLHDC2 Kelch Domain with ligand KDRLKZ-1 | | Descriptor: | GLYCEROL, Kelch domain-containing protein 2, [(5P)-5-{3-[(2R)-butan-2-yl]-7-[(2-methoxyethoxy)carbonyl]-2-oxo-5,6,7,8-tetrahydro-1,7-naphthyridin-1(2H)-yl}-2-oxopyridin-1(2H)-yl]acetic acid | | Authors: | Digianantonio, K.M, Bekes, M, Langley, D.R, Zimmerman, K. | | Deposit date: | 2023-04-12 | | Release date: | 2024-01-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.509 Å) | | Cite: | Co-opting the E3 ligase KLHDC2 for targeted protein degradation by small molecules.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8SGF
 
 | | KLHDC2 Kelch Domain with KLHDC2 c-terminal peptide bound | | Descriptor: | GLYCEROL, HIS-SER-VAL-ASN-GLN-ARG-PHE-GLY-SER-ASN-ASN-THR-SER-GLY-SER, Kelch domain-containing protein 2 | | Authors: | Digianantonio, K.M, Bekes, M, Langley, D.R, Zimmerman, K. | | Deposit date: | 2023-04-12 | | Release date: | 2024-01-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.418 Å) | | Cite: | Co-opting the E3 ligase KLHDC2 for targeted protein degradation by small molecules.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3TG6
 
 | | Crystal Structure of Influenza A Virus nucleoprotein with Ligand | | Descriptor: | Nucleocapsid protein, [4-(2-chloro-4-nitrophenyl)piperazin-1-yl][3-(2-chloropyridin-3-yl)-5-methyl-1,2-oxazol-4-yl]methanone | | Authors: | Pearce, B.C, Lewis, H.A, McDonnell, P.A, Steinbacher, S, Kiefersauer, R, Mortl, M, Maskos, K, Edavettal, S, Baldwin, E.T, Langley, D.R. | | Deposit date: | 2011-08-17 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Biophysical and Structural Characterization of a Novel Class of Influenza Virus Inhibitors
To be Published
|
|
4CL1
 
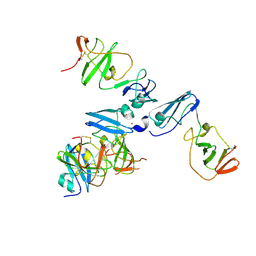 | | The crystal structure of NS5A domain 1 from genotype 1a reveals new clues to the mechanism of action for dimeric HCV inhibitors | | Descriptor: | NON-STRUCTURAL PROTEIN 5A, SULFATE ION, ZINC ION | | Authors: | Lambert, S.M, Langley, D.R, Garnett, J.A, Angell, R, Hedgethorne, K, Meanwell, N.A, Matthews, S.J. | | Deposit date: | 2014-01-10 | | Release date: | 2014-04-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The Crystal Structure of Ns5A Domain 1 from Genotype 1A Reveals New Clues to the Mechanism of Action for Dimeric Hcv Inhibitors.
Protein Sci., 23, 2014
|
|
4DYB
 
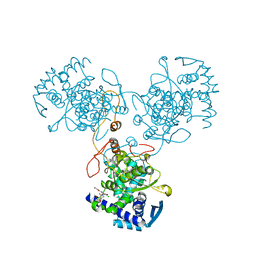 | | Crystal Structure of WSN/A Influenza Nucleoprotein with BMS-883559 Ligand Bound | | Descriptor: | N-[4-chloranyl-5-[4-[[3-(2-methoxyphenyl)-5-methyl-1,2-oxazol-4-yl]carbonyl]piperazin-1-yl]-2-nitro-phenyl]thiophene-2-carboxamide, Nucleocapsid protein | | Authors: | Lewis, H.A, Baldwin, E.T, Steinbacher, S, Maskos, K, Mortl, M, Kiefersauer, R, Edavettal, S, McDonnell, P.A, Pearce, B.C, Langley, D.R. | | Deposit date: | 2012-02-28 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | To be determined
To be Published
|
|
4DYN
 
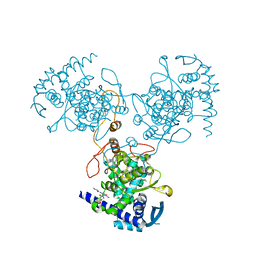 | | Crystal Structure of WSN/A Influenza Nucleoprotein with BMS-885838 Ligand Bound | | Descriptor: | N-[4-chloranyl-5-[4-[[3-(2-methoxyphenyl)-5-methyl-1,2-oxazol-4-yl]carbonyl]piperazin-1-yl]-2-nitro-phenyl]pyridine-2-carboxamide, Nucleocapsid protein | | Authors: | Lewis, H.A, Baldwin, E.T, Steinbacher, S, Maskos, K, Mortl, M, Kiefersauer, R, Edavettal, S, McDonnell, P.A, Pearce, B.C, Langley, D.R. | | Deposit date: | 2012-02-29 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | To be determined
To be Published
|
|
4DYS
 
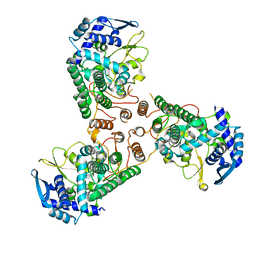 | | Crystal Structure of Apo Swine Flu Influenza Nucleoprotein | | Descriptor: | Nucleocapsid protein | | Authors: | Lewis, H.A, Baldwin, E.T, Steinbacher, S, Maskos, K, Mortl, M, Kiefersauer, R, Edavettal, S, McDonnell, P.A, Pearce, B.C, Langley, D.R. | | Deposit date: | 2012-02-29 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | To be determined
To be Published
|
|
4DYT
 
 | | Crystal Structure of WSN/A Influenza Nucleoprotein with Three Mutations (E53D, Y289H, Y313V) | | Descriptor: | Nucleocapsid protein | | Authors: | Lewis, H.A, Baldwin, E.T, Steinbacher, S, Maskos, K, Mortl, M, Kiefersauer, R, Edavettal, S, McDonnell, P.A, Pearce, B.C, Langley, D.R. | | Deposit date: | 2012-02-29 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | To be determined
To be Published
|
|
4DYA
 
 | | Crystal Structure of WSN/A Influenza Nucleoprotein with BMS-885986 Ligand Bound | | Descriptor: | N-[4-chloranyl-5-[4-[[3-(2-methoxyphenyl)-5-methyl-1,2-oxazol-4-yl]carbonyl]piperazin-1-yl]-2-nitro-phenyl]furan-2-carboxamide, Nucleocapsid protein | | Authors: | Lewis, H.A, Baldwin, E.T, Steinbacher, S, Maskos, K, Mortl, M, Kiefersauer, R, Edavettal, S, McDonnell, P.A, Pearce, B.C, Langley, D.R. | | Deposit date: | 2012-02-28 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | To be determined
To be Published
|
|
4DYP
 
 | | Crystal Structure of WSN/A Influenza Nucleoprotein with BMS-831780 Ligand Bound | | Descriptor: | Nucleocapsid protein, [4-(5-bromanyl-3-methyl-pyridin-2-yl)piperazin-1-yl]-[3-(2-chlorophenyl)-5-methyl-1,2-oxazol-4-yl]methanone | | Authors: | Lewis, H.A, Baldwin, E.T, Steinbacher, S, Maskos, K, Mortl, M, Kiefersauer, R, Edavettal, S, McDonnell, P.A, Pearce, B.C, Langley, D.R. | | Deposit date: | 2012-02-29 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | To be determined
To be Published
|
|
3RO5
 
 | | Crystal structure of influenza A virus nucleoprotein with ligand | | Descriptor: | Nucleocapsid protein, [4-(2-chloro-4-nitrophenyl)piperazin-1-yl][3-(2-methoxyphenyl)-5-methyl-1,2-oxazol-4-yl]methanone | | Authors: | Pearce, B.C, Edavettal, S, McDonnell, P.A, Lewis, H.A, Steinbacher, S, Baldwin, E.T, Langley, D.R, Maskos, K, Mortl, M, Kiefersauer, R. | | Deposit date: | 2011-04-25 | | Release date: | 2011-09-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Inhibition of influenza virus replication via small molecules that induce the formation of higher-order nucleoprotein oligomers.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6XP9
 
 | |
8SH2
 
 | | KLHDC2 in complex with EloB and EloC | | Descriptor: | Elongin-B, Elongin-C, Kelch domain-containing protein 2 | | Authors: | Digianantonio, K.M, Bekes, M. | | Deposit date: | 2023-04-13 | | Release date: | 2024-01-03 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Co-opting the E3 ligase KLHDC2 for targeted protein degradation by small molecules.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6UM8
 
 | | HIV Integrase in complex with Compound-14 | | Descriptor: | (2S)-tert-butoxy[7-(8-fluoro-5-methyl-3,4-dihydro-2H-1-benzopyran-6-yl)-5-methyl-2-phenylpyrazolo[1,5-a]pyrimidin-6-yl]acetic acid, DI(HYDROXYETHYL)ETHER, Integrase, ... | | Authors: | Khan, J.A, Kish, K. | | Deposit date: | 2019-10-09 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Discovery and Optimization of Novel Pyrazolopyrimidines as Potent and Orally Bioavailable Allosteric HIV-1 Integrase Inhibitors.
J.Med.Chem., 63, 2020
|
|
5F94
 
 | | Crystal structure of GSK3b in complex with Compound 15: 2-[(cyclopropylcarbonyl)amino]-N-(4-methoxypyridin-3-yl)pyridine-4-carboxamide | | Descriptor: | 2-[(cyclopropylcarbonyl)amino]-N-(4-methoxypyridin-3-yl)pyridine-4-carboxamide, Glycogen synthase kinase-3 beta | | Authors: | Lewis, H.A, Kish, K, Luo, G, Dubowchick, G.M. | | Deposit date: | 2015-12-09 | | Release date: | 2016-02-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery of Isonicotinamides as Highly Selective, Brain Penetrable, and Orally Active Glycogen Synthase Kinase-3 Inhibitors.
J.Med.Chem., 59, 2016
|
|
5F95
 
 | | Crystal structure of GSK3b in complex with Compound 18: 2-[(cyclopropylcarbonyl)amino]-N-(4-phenylpyridin-3-yl)pyridine-4-carboxamide | | Descriptor: | 2-[(cyclopropylcarbonyl)amino]-N-(4-phenylpyridin-3-yl)pyridine-4-carboxamide, Glycogen synthase kinase-3 beta | | Authors: | Lewis, H.A, Kish, K, Luo, G, Dubowchick, G.M. | | Deposit date: | 2015-12-09 | | Release date: | 2016-02-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.525 Å) | | Cite: | Discovery of Isonicotinamides as Highly Selective, Brain Penetrable, and Orally Active Glycogen Synthase Kinase-3 Inhibitors.
J.Med.Chem., 59, 2016
|
|
3I81
 
 | | Crystal structure of insulin-like growth factor 1 receptor (IGF-1R-WT) complex with BMS-754807 [1-(4-((5-cyclopropyl-1H-pyrazol-3-yl)amino)pyrrolo[2,1-f][1,2,4]triazin-2-yl)-N-(6-fluoro-3-pyridinyl)-2-methyl-L-prolinamide] | | Descriptor: | 1-{4-[(3-cyclopropyl-1H-pyrazol-5-yl)amino]pyrrolo[2,1-f][1,2,4]triazin-2-yl}-N-(6-fluoropyridin-3-yl)-2-methyl-L-proli namide, Insulin-like growth factor 1 receptor | | Authors: | Sack, J.S. | | Deposit date: | 2009-07-09 | | Release date: | 2009-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Discovery of a 2,4-disubstituted pyrrolo[1,2-f][1,2,4]triazine inhibitor (BMS-754807) of insulin-like growth factor receptor (IGF-1R) kinase in clinical development.
J.Med.Chem., 52, 2009
|
|
4PTE
 
 | | Structure of a carvoxamide compound (15) (N-[4-(ISOQUINOLIN-7-YL)PYRIDIN-2-YL]CYCLOPROPANECARBOXAMIDE) to GSK3b | | Descriptor: | Glycogen synthase kinase-3 beta, N-[4-(isoquinolin-7-yl)pyridin-2-yl]cyclopropanecarboxamide | | Authors: | Lewis, H.A, Sivaprakasam, P, Kish, K, Pokross, M, Dubowchik, G.M. | | Deposit date: | 2014-03-10 | | Release date: | 2015-04-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.033 Å) | | Cite: | Discovery of new acylaminopyridines as GSK-3 inhibitors by a structure guided in-depth exploration of chemical space around a pyrrolopyridinone core.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4PTC
 
 | | Structure of a carboxamide compound (3) (2-{2-[(CYCLOPROPYLCARBONYL)AMINO]PYRIDIN-4-YL}-4-OXO-4H-1LAMBDA~4~,3-THIAZOLE-5-CARBOXAMIDE) to GSK3b | | Descriptor: | 2-[2-(cyclopropylcarbonylamino)pyridin-4-yl]-4-methoxy-1,3-thiazole-5-carboxamide, Glycogen synthase kinase-3 beta | | Authors: | Lewis, H.A, Sivaprakasam, P, Kish, K, Pokross, M, Dubowchik, G.M. | | Deposit date: | 2014-03-10 | | Release date: | 2015-04-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.711 Å) | | Cite: | Discovery of new acylaminopyridines as GSK-3 inhibitors by a structure guided in-depth exploration of chemical space around a pyrrolopyridinone core.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4PTG
 
 | | Structure of a carboxamine compound (26) (2-{2-[(CYCLOPROPYLCARBONYL)AMINO]PYRIDIN-4-YL}-4-METHOXYPYRIMIDINE-5-CARBOXAMIDE) to GSK3b | | Descriptor: | 2-{2-[(cyclopropylcarbonyl)amino]pyridin-4-yl}-4-methoxypyrimidine-5-carboxamide, Glycogen synthase kinase-3 beta | | Authors: | Lewis, H.A, Sivaprakasam, P, Kish, K, Pokross, M, Dubowchik, G.M. | | Deposit date: | 2014-03-10 | | Release date: | 2015-04-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.361 Å) | | Cite: | Discovery of new acylaminopyridines as GSK-3 inhibitors by a structure guided in-depth exploration of chemical space around a pyrrolopyridinone core.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
2FXE
 
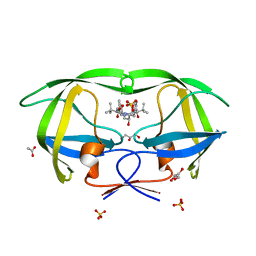 | | X-ray crystal structure of HIV-1 protease CRM mutant complexed with atazanavir (BMS-232632) | | Descriptor: | (3S,8S,9S,12S)-3,12-BIS(1,1-DIMETHYLETHYL)-8-HYDROXY-4,11-DIOXO-9-(PHENYLMETHYL)-6-[[4-(2-PYRIDINYL)PHENYL]METHYL]-2,5, 6,10,13-PENTAAZATETRADECANEDIOIC ACID DIMETHYL ESTER, ACETATE ION, ... | | Authors: | Sheriff, S, Klei, H.E. | | Deposit date: | 2006-02-05 | | Release date: | 2007-02-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structures of human immunodeficiency virus type 1 protease mutants complexed with atazanavir.
J.Virol., 81, 2007
|
|
2FXD
 
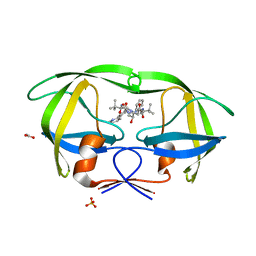 | | X-ray crystal structure of HIV-1 protease IRM mutant complexed with atazanavir (BMS-232632) | | Descriptor: | (3S,8S,9S,12S)-3,12-BIS(1,1-DIMETHYLETHYL)-8-HYDROXY-4,11-DIOXO-9-(PHENYLMETHYL)-6-[[4-(2-PYRIDINYL)PHENYL]METHYL]-2,5, 6,10,13-PENTAAZATETRADECANEDIOIC ACID DIMETHYL ESTER, ACETATE ION, ... | | Authors: | Klei, H.E, Sheriff, S. | | Deposit date: | 2006-02-04 | | Release date: | 2007-02-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystal structures of human immunodeficiency virus type 1 protease mutants complexed with atazanavir.
J.Virol., 81, 2007
|
|
2OJ9
 
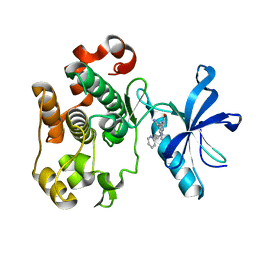 | | Structure of IGF-1R kinase domain complexed with a benzimidazole inhibitor | | Descriptor: | 3-[5-(1H-IMIDAZOL-1-YL)-7-METHYL-1H-BENZIMIDAZOL-2-YL]-4-[(PYRIDIN-2-YLMETHYL)AMINO]PYRIDIN-2(1H)-ONE, Insulin-like growth factor 1 receptor precursor (EC 2.7.10.1) (Insulin-like growth factor I receptor) (IGF-I receptor) (CD221 antigen) | | Authors: | Sack, J.S, Jacobson, B.L. | | Deposit date: | 2007-01-12 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and initial SAR of 3-(1H-benzo[d]imidazol-2-yl)pyridin-2(1H)-ones as inhibitors of insulin-like growth factor 1-receptor (IGF-1R).
Bioorg.Med.Chem.Lett., 17, 2007
|
|
5U7O
 
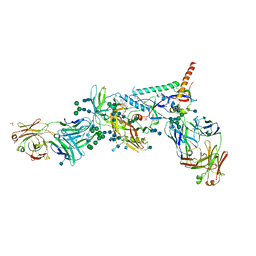 | | Crystal Structure of HIV-1 BG505 SOSIP.664 Prefusion Env Trimer Bound to Small Molecule HIV-1 Entry Inhibitor BMS-626529 in Complex with Human Antibodies PGT122 and 35O22 at 3.8 Angstrom | | Descriptor: | 1-[4-(benzenecarbonyl)piperazin-1-yl]-2-[4-methoxy-7-(3-methyl-1H-1,2,4-triazol-1-yl)-1H-pyrrolo[2,3-c]pyridin-3-yl]ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pancera, M, Lai, Y.-T, Kwong, P.D. | | Deposit date: | 2016-12-12 | | Release date: | 2017-08-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.031 Å) | | Cite: | Crystal structures of trimeric HIV envelope with entry inhibitors BMS-378806 and BMS-626529.
Nat. Chem. Biol., 13, 2017
|
|
