6UFT
 
 | |
6UL6
 
 | |
6UL4
 
 | |
6UC6
 
 | |
6UI1
 
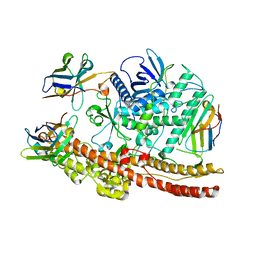 | | Crystal structure of BoNT/A-LCHn domain in complex with VHH ciA-D12, ciA-B5, and ciA-H7 | | Descriptor: | BoNT/A, ciA-B5, ciA-D12, ... | | Authors: | Lam, K, Jin, R. | | Deposit date: | 2019-09-29 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.20000863 Å) | | Cite: | Structural Insights into Rational Design of Single-Domain Antibody-Based Antitoxins against Botulinum Neurotoxins
Cell Rep, 30, 2020
|
|
6UHT
 
 | |
3QG5
 
 | |
4I51
 
 | | Methyltransferase domain of HUMAN EUCHROMATIC HISTONE METHYLTRANSFERASE 1, mutant Y1211A | | Descriptor: | GLYCEROL, H3K9 NE-ALLYL PEPTIDE, Histone-lysine N-methyltransferase EHMT1, ... | | Authors: | Dong, A, Zeng, H, Walker, J.R, Islam, K, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Lou, M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-11-28 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Defining efficient enzyme-cofactor pairs for bioorthogonal profiling of protein methylation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7ZQY
 
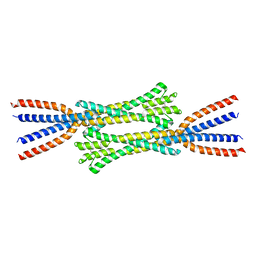 | | Chaetomium thermophilum Rad50 Zn hook | | Descriptor: | DH domain-containing protein, ZINC ION | | Authors: | Lammens, K, Rotheneder, M, Stakyte, K. | | Deposit date: | 2022-05-03 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Cryo-EM structure of the Mre11-Rad50-Nbs1 complex reveals the molecular mechanism of scaffolding functions.
Mol.Cell, 83, 2023
|
|
3QF7
 
 | |
7PPJ
 
 | | human SLFN5 | | Descriptor: | Schlafen family member 5, ZINC ION | | Authors: | Lammens, K, Metzner, F.J. | | Deposit date: | 2021-09-14 | | Release date: | 2022-01-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structural and biochemical characterization of human Schlafen 5.
Nucleic Acids Res., 50, 2022
|
|
8S86
 
 | | human PLD3 homodimer structure | | Descriptor: | 5'-3' exonuclease PLD3 | | Authors: | Lammens, K. | | Deposit date: | 2024-03-05 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Lysosomal endonuclease RNase T2 and PLD exonucleases cooperatively generate RNA ligands for TLR7 activation.
Immunity, 57, 2024
|
|
3KOR
 
 | | Crystal structure of a putative Trp repressor from Staphylococcus aureus | | Descriptor: | Possible Trp repressor | | Authors: | Lam, R, Vodsedalek, J, Lam, K, Romanov, V, Battaile, K.P, Beletskaya, I, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2009-11-13 | | Release date: | 2010-11-17 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a putative Trp repressor from Staphylococcus aureus
To be Published
|
|
3L5A
 
 | | Crystal structure of a probable NADH-dependent flavin oxidoreductase from Staphylococcus aureus | | Descriptor: | NADH/flavin oxidoreductase/NADH oxidase, TRIETHYLENE GLYCOL | | Authors: | Lam, R, Gordon, R.D, Vodsedalek, J, Battaile, K.P, Grebemeskel, S, Lam, K, Romanov, V, Chan, T, Mihajlovic, V, Thompson, C.M, Guthrie, J, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2009-12-21 | | Release date: | 2010-12-22 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a probable NADH-dependent flavin oxidoreductase from Staphylococcus aureus
To be Published
|
|
3KWL
 
 | | Crystal structure of a hypothetical protein from Helicobacter pylori | | Descriptor: | uncharacterized protein | | Authors: | Lam, R, Thompson, C.M, Vodsedalek, J, Lam, K, Romanov, V, Battaile, K.P, Beletskaya, I, Gordon, E, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2009-12-01 | | Release date: | 2010-12-01 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of a hypothetical protein from Helicobacter pylori
To be Published
|
|
3E2I
 
 | | Crystal structure of Thymidine Kinase from S. aureus | | Descriptor: | GLYCEROL, Thymidine kinase, ZINC ION | | Authors: | Lam, R, Johns, K, Battaile, K.P, Romanov, V, Lam, K, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2008-08-05 | | Release date: | 2009-08-11 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of Thymidine Kinase from S. aureus
To be Published
|
|
3D7R
 
 | | Crystal structure of a putative esterase from Staphylococcus aureus | | Descriptor: | BROMIDE ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Lam, R, Battaile, K.P, Chan, T, Romanov, V, Lam, K, Soloveychik, M, Wu-Brown, J, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2008-05-21 | | Release date: | 2009-06-09 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of a putative esterase from Staphylococcus aureus
To be Published
|
|
4H1O
 
 | | Crystal structure of the tyrosine phosphatase SHP-2 with D61G mutation | | Descriptor: | 1,2-ETHANEDIOL, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Qiu, W, Lin, A, Hutchinson, A, Romanov, V, Ruzanov, M, Thompson, C, Lam, K, Kisselman, G, Battaile, K, Chirgadze, N.Y. | | Deposit date: | 2012-09-11 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the tyrosine phosphatase SHP-2 with D61G mutation
To be Published
|
|
4H34
 
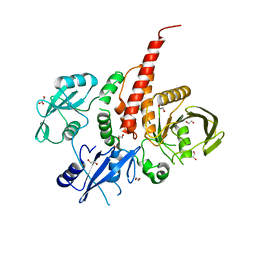 | | Crystal structure of the tyrosine phosphatase SHP-2 with Q506P mutation | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Qiu, W, Lin, A, Hutchinson, A, Romanov, V, Ruzanov, M, Thompson, C, Lam, K, Kisselman, G, Battaile, K, Chirgadze, N.Y. | | Deposit date: | 2012-09-13 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the tyrosine phosphatase SHP-2 with Q506P mutation
To be Published
|
|
4GWF
 
 | | Crystal structure of the tyrosine phosphatase SHP-2 with Y279C mutation | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Qiu, W, Lin, A, Hutchinson, A, Romanov, V, Ruzanov, M, Thompson, C, Lam, K, Kisselman, G, Battaile, K, Chirgadze, N.Y. | | Deposit date: | 2012-09-03 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the tyrosine phosphatase SHP-2 with Y279C mutation
TO BE PUBLISHED
|
|
4PAW
 
 | | Structure of hypothetical protein HP1257. | | Descriptor: | Orotate phosphoribosyltransferase, PHOSPHATE ION, THIOCYANATE ION, ... | | Authors: | Battaile, K.P, Lam, R, Lam, K, Romanov, V, Jones, K, Soloveychik, M, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-04-10 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure of hypothetical protein HP1257.
To Be Published
|
|
4PAV
 
 | | Structure of hypothetical protein SA1046 from S. aureus. | | Descriptor: | Glyoxalase family protein | | Authors: | Battaile, K.P, Mulichak, A, Lam, R, Lam, K, Soloveychik, M, Romanov, V, Jones, K, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-04-10 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of hypothetical protein SA1046 from S. aureus.
To Be Published
|
|
3K2A
 
 | | Crystal structure of the homeobox domain of human homeobox protein Meis2 | | Descriptor: | ACETATE ION, CHLORIDE ION, Homeobox protein Meis2 | | Authors: | Lam, R, Soloveychik, M, Battaile, K.P, Romanov, V, Lam, K, Beletskaya, I, Gordon, E, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2009-09-29 | | Release date: | 2010-10-13 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the homeobox domain of human homeobox protein Meis2
To be Published
|
|
4NWF
 
 | | Crystal structure of the tyrosine phosphatase SHP-2 with N308D mutation | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Qiu, W, Lin, A, Hutchinson, A, Romanov, V, Ruzanov, M, Thompson, C, Lam, K, Kisselman, G, Battalie, K, Chirgadze, N.Y. | | Deposit date: | 2013-12-06 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the tyrosine phosphatase SHP-2 with N308D mutation
To be Published
|
|
4NWG
 
 | | Crystal structure of the tyrosine phosphatase SHP-2 with E139D mutation | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Qiu, W, Lin, A, Hutchinson, A, Romanov, V, Ruzanov, M, Thompson, C, Lam, K, Kisselman, G, Battalie, K, Chirgadze, N.Y. | | Deposit date: | 2013-12-06 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of the tyrosine phosphatase SHP-2 with E139D mutation
To be Published
|
|
