4V4B
 
 | | Structure of the ribosomal 80S-eEF2-sordarin complex from yeast obtained by docking atomic models for RNA and protein components into a 11.7 A cryo-EM map. | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S11, ... | | Authors: | Spahn, C.M, Gomez-Lorenzo, M.G, Grassucci, R.A, Jorgensen, R, Andersen, G.R, Beckmann, R, Penczek, P.A, Ballesta, J.P.G, Frank, J. | | Deposit date: | 2004-01-06 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (11.7 Å) | | Cite: | Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation.
Embo J., 23, 2004
|
|
4US2
 
 | | The crystal structure of H-Ras and SOS in complex with ligands | | Descriptor: | 3-[(3R)-1-ethyl-2,5-dioxopyrrolidin-3-yl]benzamide, GTPASE HRAS, SON OF SEVENLESS HOMOLOG 1 | | Authors: | Winter, J.J.G, Anderson, M, Blades, K, Brassington, C, Breeze, A.L, Chresta, C, Embrey, K, Fairley, G, Faulder, P, Finlay, M.R.V, Kettle, J.G, Nowak, T, Overman, R, Patel, S.J, Perkins, P, Spadola, L, Tart, J, Tucker, J, Wrigley, G. | | Deposit date: | 2014-07-02 | | Release date: | 2015-03-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Small Molecule Binding Sites on the Ras:SOS Complex Can be Exploited for Inhibition of Ras Activation.
J.Med.Chem., 58, 2015
|
|
4UCR
 
 | | Fragment bound to H.influenza NAD dependent DNA ligase | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 8-hydroxy-2-methylquinoline-6-carboxamide, DNA LIGASE | | Authors: | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
4UCO
 
 | | Fragment bound to H.influenza NAD dependent DNA ligase | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 7-amino-2-tert-butyl-4-(1H-pyrrol-2-yl)pyrido[2,3-d]pyrimidine-6-carboxamide, DNA LIGASE | | Authors: | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
4UCS
 
 | | Fragment bound to H.influenza NAD dependent DNA ligase | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 5-amino-3-(furan-2-yl)-1H-1,2,4-triazole-1-carboxamide, DNA LIGASE | | Authors: | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
4UTQ
 
 | | A structural model of the active ribosome-bound membrane protein insertase YidC | | Descriptor: | ATP SYNTHASE SUBUNIT C, MEMBRANE PROTEIN INSERTASE YIDC | | Authors: | Wickles, S, Singharoy, A, Andreani, J, Seemayer, S, Bischoff, L, Berninghausen, O, Soeding, J, Schulten, K, vanderSluis, E.O, Beckmann, R. | | Deposit date: | 2014-07-22 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | A Structural Model of the Active Ribosome-Bound Membrane Protein Insertase Yidc.
Elife, 3, 2014
|
|
4V26
 
 | | VER-246608, a novel pan-isoform ATP competitive inhibitor of pyruvate dehydrogenase kinase, disrupts Warburg metabolism and induces context- dependent cytostasis in cancer cells | | Descriptor: | MAGNESIUM ION, N-(2-AMINOETHYL)-2-{3-CHLORO-4-[(4-ISOPROPYLBENZYL)OXY]PHENYL} ACETAMIDE, N-[4-(2-CHLORO-5-METHYLPYRIMIDIN-4-YL)PHENYL]-2,4-DIHYDROXY-N-(4-{[(TRIFLUOROACETYL)AMINO]METHYL}BENZYL)BENZAMIDE, ... | | Authors: | Moore, J.D, Staniszewska, A, Shaw, T, D'Alessandro, J, Davis, B, Surgenor, A, Baker, L, Matassova, N, Murray, J, Macias, A, Brough, P, Wood, M, Mahon, P.C. | | Deposit date: | 2014-10-06 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | VER-246608, a novel pan-isoform ATP competitive inhibitor of pyruvate dehydrogenase kinase, disrupts Warburg metabolism and induces context-dependent cytostasis in cancer cells.
Oncotarget, 5, 2014
|
|
4UF0
 
 | | Crystal structure of JmjC domain of human histone demethylase UTY in complex with epitherapuetic compound 2-(((2-((2-(dimethylamino)ethyl) (ethyl)amino)-2-oxoethyl)amino)methyl)isonicotinic acid. | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(2-{[(E)-2-(dimethylamino)ethenyl](ethyl)amino}-2-oxoethyl)amino]methyl}pyridine-4-carboxylic acid, FE (II) ION, ... | | Authors: | Srikannathasan, V, Johansson, C, Gileadi, C, Tobias, K, Kopec, J, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2014-12-22 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
4UVK
 
 | |
4UFZ
 
 | | Synthesis of Novel NAD Dependant DNA Ligase Inhibitors via Negishi Cross-Coupling: Development of SAR and Resistance Studies | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 5,7-bis(azanyl)-2-tert-butyl-4-(1,3-thiazol-2-yl)pyrido[2,3-d]pyrimidine-6-carbonitrile, DNA LIGASE | | Authors: | Murphy-Benenato, K.E, Boriack-Sjodin, P.A, Martinez-Botella, G, Carcanague, D, Gingipali, L, Gowravaram, M, Harang, J, Hale, M, Ioannidis, G, Jahic, H, Johnstone, M, Kutschke, A, Laganas, V.A, Loch, J, Oguto, H, Patel, S.J. | | Deposit date: | 2015-03-20 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Negishi Cross-Coupling Enabled Synthesis of Novel Nad(+)-Dependent DNA Ligase Inhibitors and Sar Development.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
7M78
 
 | | Room Temperature XFEL Crystallography reveals asymmetry in the vicinity of the two phylloquinones in Photosystem I | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Keable, S.M, Simon, P.S, Kolsch, A, Kern, J, Yachandra, V.K, Zouni, A, Yano, J. | | Deposit date: | 2021-03-26 | | Release date: | 2021-11-24 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Room temperature XFEL crystallography reveals asymmetry in the vicinity of the two phylloquinones in photosystem I.
Sci Rep, 11, 2021
|
|
7M76
 
 | | Room Temperature XFEL Crystallography reveals asymmetry in the vicinity of the two phylloquinones in Photosystem I | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Keable, S.M, Simon, P.S, Kolsch, A, Kern, J, Yachandra, V.K, Zouni, A, Yano, J. | | Deposit date: | 2021-03-26 | | Release date: | 2021-11-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Room temperature XFEL crystallography reveals asymmetry in the vicinity of the two phylloquinones in photosystem I.
Sci Rep, 11, 2021
|
|
7M75
 
 | | Room Temperature XFEL Crystallography reveals asymmetry in the vicinity of the two phylloquinones in Photosystem I | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Keable, S.M, Simon, P.S, Kolsch, A, Kern, J, Yachandra, V.K, Zouni, A, Yano, J. | | Deposit date: | 2021-03-26 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Room temperature XFEL crystallography reveals asymmetry in the vicinity of the two phylloquinones in photosystem I.
Sci Rep, 11, 2021
|
|
4TPI
 
 | | THE REFINED 2.2-ANGSTROMS (0.22-NM) X-RAY CRYSTAL STRUCTURE OF THE TERNARY COMPLEX FORMED BY BOVINE TRYPSINOGEN, VALINE-VALINE AND THE ARG15 ANALOGUE OF BOVINE PANCREATIC TRYPSIN INHIBITOR | | Descriptor: | BOVINE PANCREATIC TRYPSIN INHIBITOR, CALCIUM ION, SULFATE ION, ... | | Authors: | Bode, W, Walter, J. | | Deposit date: | 1985-06-11 | | Release date: | 1985-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The refined 2.2-A (0.22-nm) X-ray crystal structure of the ternary complex formed by bovine trypsinogen, valine-valine and the Arg15 analogue of bovine pancreatic trypsin inhibitor
Eur.J.Biochem., 144, 1984
|
|
9DW4
 
 | | Dephosphorylated CFTR in 1:1 complex with PKA-C (site II) | | Descriptor: | Cystic fibrosis transmembrane conductance regulator, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Fiedorczuk, K, Chen, J, Csanady, L. | | Deposit date: | 2024-10-08 | | Release date: | 2024-11-13 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | The structures of protein kinase A in complex with CFTR: Mechanisms of phosphorylation and noncatalytic activation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9DW5
 
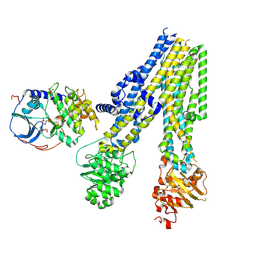 | | Dephosphorylated CFTR in 1:1 complex with PKA-C (site I) | | Descriptor: | Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Fiedorczuk, K, Chen, J, Csanady, L. | | Deposit date: | 2024-10-08 | | Release date: | 2024-11-13 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The structures of protein kinase A in complex with CFTR: Mechanisms of phosphorylation and noncatalytic activation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9DW7
 
 | | Dephosphorylated CFTR in 1:2 complex with PKA-C | | Descriptor: | Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Fiedorczuk, K, Chen, J, Csanady, L. | | Deposit date: | 2024-10-08 | | Release date: | 2024-11-13 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | The structures of protein kinase A in complex with CFTR: Mechanisms of phosphorylation and noncatalytic activation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9DW9
 
 | | Phosphorylated (E1371Q)CFTR in complex with PKA-C | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, CHOLESTEROL, ... | | Authors: | Fiedorczuk, K, Chen, J, Csanady, L. | | Deposit date: | 2024-10-08 | | Release date: | 2024-11-13 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The structures of protein kinase A in complex with CFTR: Mechanisms of phosphorylation and noncatalytic activation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9DW8
 
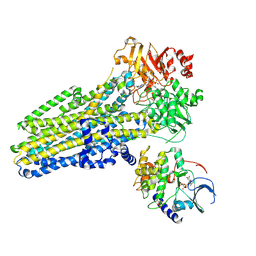 | | Dephosphorylated (E1371Q)CFTR in complex with PKA-C | | Descriptor: | Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION, N-(2-phenylethyl)adenosine 5'-(tetrahydrogen triphosphate), ... | | Authors: | Fiedorczuk, K, Chen, J, Csanady, L. | | Deposit date: | 2024-10-08 | | Release date: | 2024-11-13 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The structures of protein kinase A in complex with CFTR: Mechanisms of phosphorylation and noncatalytic activation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7M72
 
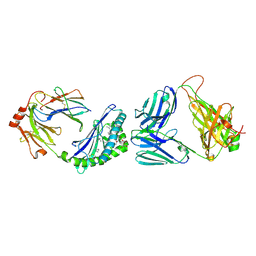 | | MHC-like protein complex structure | | Descriptor: | (3R)-N-[(2S,3R)-1-(alpha-D-galactopyranosyloxy)-3-hydroxy-15-methylhexadecan-2-yl]-3-hydroxyheptadecanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Thirunavukkarasu, P, Le Nours, J, Rossjohn, J. | | Deposit date: | 2021-03-26 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Host immunomodulatory lipids created by symbionts from dietary amino acids.
Nature, 600, 2021
|
|
6YVF
 
 | | Structure of SARS-CoV-2 Main Protease bound to AZD6482. | | Descriptor: | 2-[[(1R)-1-(7-methyl-2-morpholin-4-yl-4-oxidanylidene-pyrido[1,2-a]pyrimidin-9-yl)ethyl]amino]benzoic acid, 3C-like proteinase, CALCIUM ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6DHH
 
 | | RT XFEL structure of Photosystem II 400 microseconds after the second illumination at 2.2 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Chatterjee, R, Young, I.D, Fuller, F.D, Lassalle, L, Ibrahim, M, Gul, S, Fransson, T, Brewster, A.S, Alonso-Mori, R, Hussein, R, Zhang, M, Douthit, L, de Lichtenberg, C, Cheah, M.H, Shevela, D, Wersig, J, Seufert, I, Sokaras, D, Pastor, E, Weninger, C, Kroll, T, Sierra, R.G, Aller, P, Butryn, A, Orville, A.M, Liang, M, Batyuk, A, Koglin, J.E, Carbajo, S, Boutet, S, Moriarty, N.W, Holton, J.M, Dobbek, H, Adams, P.D, Bergmann, U, Sauter, N.K, Zouni, A, Messinger, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2018-05-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of the intermediates of Kok's photosynthetic water oxidation clock.
Nature, 563, 2018
|
|
3CG8
 
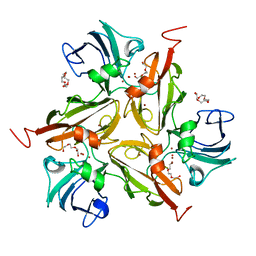 | | Laccase from Streptomyces coelicolor | | Descriptor: | COPPER (II) ION, OXYGEN ATOM, TETRAETHYLENE GLYCOL, ... | | Authors: | Skalova, T, Dohnalek, J, Ostergaard, L.H, Ostergaard, P.R, Kolenko, P, Duskova, J, Hasek, J. | | Deposit date: | 2008-03-05 | | Release date: | 2009-01-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.679 Å) | | Cite: | The Structure of the Small Laccase from Streptomyces coelicolor Reveals a Link between Laccases and Nitrite Reductases.
J.Mol.Biol., 385, 2009
|
|
6DHF
 
 | | RT XFEL structure of the one-flash state of Photosystem II (1F, S2-rich) at 2.08 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Chatterjee, R, Young, I.D, Fuller, F.D, Lassalle, L, Ibrahim, M, Gul, S, Fransson, T, Brewster, A.S, Alonso-Mori, R, Hussein, R, Zhang, M, Douthit, L, de Lichtenberg, C, Cheah, M.H, Shevela, D, Wersig, J, Seufert, I, Sokaras, D, Pastor, E, Weninger, C, Kroll, T, Sierra, R.G, Aller, P, Butryn, A, Orville, A.M, Liang, M, Batyuk, A, Koglin, J.E, Carbajo, S, Boutet, S, Moriarty, N.W, Holton, J.M, Dobbek, H, Adams, P.D, Bergmann, U, Sauter, N.K, Zouni, A, Messinger, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2018-05-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structures of the intermediates of Kok's photosynthetic water oxidation clock.
Nature, 563, 2018
|
|
1D07
 
 | | Hydrolytic haloalkane dehalogenase linb from sphingomonas paucimobilis UT26 with 1,3-propanediol, a product of debromidation of dibrompropane, at 2.0A resolution | | Descriptor: | 1,3-PROPANDIOL, BROMIDE ION, HALOALKANE DEHALOGENASE | | Authors: | Marek, J, Vevodova, J, Damborsky, J, Smatanova, I, Svensson, L.A, Newman, J, Nagata, Y, Takagi, M. | | Deposit date: | 1999-09-09 | | Release date: | 2000-09-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the haloalkane dehalogenase from Sphingomonas paucimobilis UT26.
Biochemistry, 39, 2000
|
|
